Mutation update for CYP4F22 variants associated with autosomal recessive congenital ichthyosis
Funding information:
This study was supported by a grant from the German research foundation DFG (FI1767/3-1).
Communicated by Johannes Zschocke
Abstract
Autosomal recessive congenital ichthyosis (ARCI) is a heterogeneous group of rare disorders of keratinization characterized by generalized abnormal scaling of the skin. Ten genes are currently known to be associated with ARCI: TGM1, ALOXE3, ALOX12B, NIPAL4 (ICHTHYIN), ABCA12, CYP4F22, PNPLA1, CERS3, SDR9C7, and SULT2B1. Over a period of 22 years, we have studied a large patient cohort from 770 families with a clinical diagnosis of ARCI. Since the first report that mutations in the gene CYP4F22 are causative for ARCI in 2006, we have identified 54 families with pathogenic mutations in CYP4F22 including 23 previously unreported mutations. In this report, we provide an up-to-date overview of all published and novel CYP4F22 mutations and point out possible mutation hot spots. We discuss the molecular and clinical findings, the genotype–phenotype correlations and consequences on genetic testing.
1 INTRODUCTION
Autosomal recessive congenital ichthyosis (ARCI) is a heterogeneous group of rare disorders of keratinization characterized by generalized abnormal scaling of the skin. The main skin phenotypes of ARCI are lamellar ichthyosis (LI), congenital ichthyosiform erythroderma (CIE), and harlequin ichthyosis (HI). Minor variants of ARCI are self-improving collodion ichthyosis (SICI, acronym coined by Vahlquist et al., 2010) and bathing suit ichthyosis (BSI). Ten genes have been identified to be causative for ARCI: TGM1, ALOXE3, ALOX12B, NIPAL4 (ICHTHYIN), ABCA12, CYP4F22, PNPLA1, CERS3, SDR9C7, and SULT2B1. Mutations in LIPN have also been classified as ARCI, but do not lead to a congenital form of ichthyosis.
CYP4F22 (MIM* 611495) is located on chromosome 19p13.12. CYP4F22 is a protein of the cytochrome-P450 family 4 and encodes an epidermal ω-hydroxylase important for the synthesis of ω-hydroxyceramides in the epidermis, which is hypothesized to be crucial for skin-barrier function (Behne et al., 2000). For a long time, little was known about the function of CYP4F22 in the metabolic pathway in the skin. Functional studies identified CYP4F22 as the long-sought fatty acid ω-hydroxylase (Ohno et al., 2015), which catalyzes the ω-hydroxylation of ultra-long-chain (ULC) fatty acids in the pathway of acylceramide production. CYP4F22 is localized in the membrane of the endoplasmic reticulum (Ohno et al., 2015). The ω-hydroxylation occurs on the cytoplasmic side of the endoplasmic reticulum (Akiyama, 2017). Studies of Ohno et al. (2015) showed that mutant proteins exhibit reduced enzyme activity, which suggests a correlation between activity and pathology. Lipid analysis of a patient with ichthyosis carrying CYP4F22 mutations showed a decrease in acylceramide production (Ohno et al., 2015).
In this mutation update, we review the current state of our knowledge of CYP4F22 mutations and present 23 previously unpublished mutations. We examine the molecular and clinical findings, discuss the genotype–phenotype correlation and compare our findings to the literature.
2 METHODS
Our ARCI cohort includes 54 families with CYP4F22 mutations. In this study, we present one affected member from each family. Informed consent was obtained from all patients. This study was conducted according to the Declaration of Helsinki principles. Genomic DNA was isolated from peripheral blood leucocytes by standard methods. Mutation screenings were performed using standard methods for PCR amplification, Sanger sequencing and next generation sequencing (NGS). For mutation analysis, all coding exons and adjacent splice sites of the CYP4F22 gene were PCR amplified using exon-specific primer pairs, which were established in our laboratory. Primer sequences can be provided on request. Automated DNA sequence analysis was performed using an ABI 3500 DNA Sequencer (Applied Biosystems, Foster City, USA). In four patients, we applied NGS methods through multi-gene panel testing. We used a HaloPlex Custom Kit (Agilent Technologies, Inc. Santa Clara, CA, USA) for enrichment of the sequences and performed sequence analysis using the Illumina MiSeq instrument (Illumina, San Diego, CA, USA). Bioinformatics analysis was done using the commercial software SeqNext (JSI medical systems) and an in-house bioinformatics pipeline. All mutations were confirmed by a second PCR with an independent DNA extraction.
CYP4F22 variants are described according to current HGVS mutation nomenclature guidelines (https://varnomen.hgvs.org; den Dunnen et al., 2016) ascribing the A of the first ATG translational initiation codon as nucleotide + 1. We used the CYP4F22 reference sequence NM_173483.3. The CYP4F22 gene contains 14 exons and encodes 531 amino acids. The first two exons of CYP4F22 are non-coding and the first ATG translational initiation codon is located in exon 3.
The following databases were used for this study: The Exome Aggregation Consortium (ExAC; https://exac.broadinstitute.org/; Lek et al., 2016), The Genome Aggregation Database (gnomAD; https://gnomad.broadinstitute.org/), Database of Single-Nucleotide Polymorphisms (dbSNP; https://www.ncbi.nlm.nih.gov/projects/SNP/), HGMD Professional (https://www.biobase-international.com/product/hgmd), PubMed (https://www.ncbi.nlm.nih.gov/pubmed/).
For the prediction of the mutation effect, we used the following bioinformatic tools (Supp. Table S1): Mutation Taster (https://www.mutationtaster.org/; Schwarz, Cooper, Schuelke, & Seelow, 2014), PolyPhen-2 (https://genetics.bwh.harvard.edu/pph2/, Adzhubei et al., 2010), SIFT (https://sift.jcvi.org/; Ng & Henikoff, 2001), Provean (https://provean.jcvi.org/index.php; Choi & Chan, 2015), fathmm (https://fathmm.biocompute.org.uk/; Shihab et al., 2014), Human Splicing Finder version 3.0 (https://www.umd.be/HSF3/; Desmet et al., 2009), NetGene2 (https://www.cbs.dtu.dk/services/NetGene2/; Hebsgaard et al., 1996), NNSplice version 0.9 (https://www.fruitfly.org/; Reese, Eeckman, Kulp, & Haussler, 1997).
For morphological examination, skin samples of patients P52 and P53 were cut in 1 mm3 samples and fixed in 2.5% glutaraldehyde (P52) or 4% paraformaldehyde/2% glutaraldehyde (P53), postfixed in 1% osmium tetroxide solution for 1 h, dehydrated in ethanol (25, 50, 75, 90, 100%) and propylene oxide and embedded in an epoxy resin. Sections (70 nm) were mounted on copper grids, stained with 5% uranyl acetate and Reynold's solution and examined in a Philips EM400.
3 THE SPECTRUM OF CYP4F22 MUTATIONS
We studied patients with a clinical diagnosis of ARCI from a large cohort of 770 families. The affected individuals presented ichthyosis symptoms at birth and no signs of systemic disease. For mutation analysis, we performed Sanger sequencing of ARCI genes and/or next generation sequencing with multi gene panels. In 54 families, we detected mutations in the CYP4F22 gene. We present 23 previously unreported pathogenic mutations. Thirteen patients of our cohort have previously been reported in Lefevre et al. (2006) and Pigg et al. (2016) (see Table 1). Before this report, 21 different pathogenic mutations in CYP4F22 had been reported in patients affected with ARCI (Bučková et al., 2016, Diociaiuti et al., 2016, Gruber et al., 2017, Israeli et al., 2013, Lefèvre et al., 2006, Lugassy et al., 2008, Palamar et al., 2015, Pigg et al., 2016, Scott et al. 2013, Sugiura et al., 2013). Taken together, the total number of pathogenic mutations known to date is 44 (Table 1, Figure 1, and Supp. Table S1). All mutations identified by our study and the literature review were submitted to the database ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/). The types of mutations include missense mutations (26 out of 44; 59.1%), frameshift mutations (7 out of 44; 15.9%), splice-site mutations (6 out of 44; 13.6%), nonsense mutations (4 out of 44; 9.1%) and a large deletion (1 out of 44; 2.3%). Most of these mutations occur only in one family. The most frequently detected mutation is p.His435Tyr, which was found in 15 families. Except two patients from Germany and Cap Verde, all other families with this mutation come from the same area of Algeria. Some amino acid positions seem to be mutation hot spots. The mutation p.Arg156Cys is a recurrent mutation, which was found in three families of different origins. In addition, we detected another pathogenic amino acid change at this position, p.Arg156His. A second mutation hotspot is at amino acid position 243. In eight families from different origins, the mutation p.Arg243His was detected, and in one family, the mutation p.Arg243Cys was found. Further potential mutation hotspots are at amino acid position 362 (p.Arg362Gln, p.Arg362Gly, and p.Arg362*) and 451 (p.Arg451Cys and p.Arg451Pro). The most frequent frameshift mutation is c.59dup which was found in six families from different origins. Buckòvá et al. (2016) postulated the existence of a founder effect for this mutation, since both Czech families analyzed in their study are distantly related. However, four families from our cohort come from Germany, Switzerland, and Denmark.
| Family | Exon/Intron | Nucleotide change | Amino acid change | Zygosity | Type | Reference | Comment |
|---|---|---|---|---|---|---|---|
| 1 | 3 | c.177C > G | p.Phe59Leu | hom | Missense | Lefèvre et al. (2006) | 1 |
| 2 | 3 | c.177C > G | p.Phe59Leu | hom | Missense | Lefèvre et al. (2006) | |
| 3 | 3 | c.177C > G | p.Phe59Leu | hom | Missense | Lefèvre et al. (2006) | |
| 4 | 4 | c.242G > A | p.Gly81Asp | hom | Missense | This study | |
| 5 | 6 | c.493_499delinsCTTGATT | p.165_167delinsLeuAspPhe | hom | Indel | This study | |
| 6 | 6 | c.550-2A > T | p.spl? | hom | Splice | This study | |
| 7 | 7 | c.592G > T | p.Asp198Tyr | hom | Missense | This study | |
| 8 | 7 | c.643_644del | p.Val215Leufs*13 | hom | Frameshift | This study | |
| 9 | 7 | c.667C > T | p.Gln223* | hom | Nonsense | Pigg et al. (2016) | 2 |
| 10 | 8 | c.728G > A | p.Arg243His | hom | Missense | Lefèvre et al. (2006) | |
| 11 | 8 | c.728G > A | p.Arg243His | hom | Missense | Lefèvre et al. (2006) | |
| 12 | 8 | c.728G > A | p.Arg243His | hom | Missense | Lefèvre et al. (2006) | 1 |
| 13 | 8 | c.728G > A | p.Arg243His | hom | Missense | Lefèvre et al. (2006) | |
| 14 | 8 | c.847C > T | p.Arg283Trp | hom | Missense | This study | |
| 15 | 8 | c.917T > C | p.Ile306Thr | hom | Missense | This study | |
| 16 | 9 | c.976C > T | p.Arg326* | hom | Stopgain | Palamar et al. (2015) | |
| 17 | 9 | c.981del | p.Glu328Lysfs*42 | hom | Frameshift | Lefèvre et al. (2006) | |
| 18 | 9 | c.981del | p.Glu328Lysfs*42 | hom | Frameshift | Lefèvre et al. (2006) | 1 |
| 19 | 9 | c.981del | p.Glu328Lysfs*42 | hom | Frameshift | Lefèvre et al. (2006) | |
| 20 | 10 | c.1084C > G | p.Arg362Gly | hom | Missense | This study | |
| 21 | 10 | c.1114C > T | p.Arg372Trp | hom | Missense | Lefèvre et al. (2006) | 1 |
| 22 | 10 | c.1114C > T | p.Arg372Trp | hom | Missense | Lefèvre et al. (2006) | |
| 23 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | |
| 24 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | 1 |
| 25 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | |
| 26 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | |
| 27 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | 1 |
| 28 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | 1 |
| 29 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | 1 |
| 30 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | 1 |
| 31 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | |
| 32 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | |
| 33 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | |
| 34 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | |
| 35 | 12 | c.1303C > T | p.His435Tyr | hom | Missense | Lefèvre et al. (2006) | |
| 36 | 12 | c.1306C > G | p.His436Asp | hom | Missense | Lefèvre et al. (2006) | 1 |
| 37 | 13 | c.1351C > T | p.Arg451Cys | hom | Missense | This study | |
| 38 | 13 | c.1351C > T | p.Arg451Cys | hom | Missense | This study | |
| 39 | 14 | c.1488C > G | p.Phe496Leu | hom | Missense | This study | |
| 40 | 14 | c.1532A > G | p.Glu511Gly | hom | Missense | This study | |
| 41 | 5-11 | c.368-?_1596+?del (deletion of exon 5–14) | p.spl? | hom | Deletion | Lefèvre et al. (2006) | 1 |
| 42 | 3 | c.59dup | p.Ile21Hisfs*59 | het | Frameshift | Bučková et al. (2016) | |
| 4 | c.314C > T | p.Pro105Leu | het | Missense | This study | ||
| 43 | 3 | c.59dup | p.Ile21Hisfs*58 | het | Frameshift | Bučková et al. (2016) | |
| 8 | c.720_723del | p.Val241Glyfs*128 | het | Frameshift | This study | ||
| 44 | 3 | c.59dup | p.Ile21Hisfs*59 | het | Frameshift | Bučková et al. (2016) | 2 |
| 8 | c.727C > T | p.Arg243Cys | het | Missense | Pigg et al. (2016) | ||
| 45 | 3 | c.59dup | p.Ile21Hisfs*59 | het | Frameshift | Bučková et al. (2016) | |
| 12 | c.1303C > T | p.His435Tyr | het | Missense | Lefèvre et al. (2006) | ||
| 46 | 3 | c.1352G > C | p.Arg451Pro | het | Missense | This study | |
| 14 | c.1488C > G | p.Phe496Leu | het | Missense | This study | ||
| 47 | 4 | c.242G > A | p.Gly81Asp | het | Missense | This study | |
| 8 | c.940-1G > A | p.spl? | het | Splice | This study | ||
| 48 | 5 | c.421+1G > A | p.spl? | het | Splice | This study | |
| 10 | c.1064C > T | p.Pro355Leu | het | Missense | This study | ||
| 49 | 6 | c.466C > T | p.Arg156Cys | het | Missense | Israeli et al. (2013) | |
| 6 | c.467G > A | p.Arg156His | het | Missense | This study | ||
| 50 | 8 | c.728G > A | p.Arg243His | het | Missense | Lefèvre et al. (2006) | |
| 10 | c.1084C > T | p.Arg362* | het | Nonsense | Diociauti et al. (2006) | ||
| 51 | 8 | c.697A > C | p.Ile233Leu | het | Missense | This study | |
| 11 | c.1163C > A | p.Thr388Lys | het | Missense | This study | ||
| 52 | 8 | c.912C > A | p.Asp304Glu | het | Missense | This study | |
| 14 | c.1424G > A | p.Cys475Tyr | het | Missense | This study | ||
| 53 | 9 | c.976C > T | p.Arg326* | het | Stopgain | Palamar et al. (2015) | |
| 10 | c.1137-18_1137-4del | p.spl? | het | Splice | This study | ||
| 54 | 9 | c.976C > T | p.Arg326* | het | Nonsense | Palamar et al. (2015) | |
| 12 | c.1303C > T | p.His435Tyr | het | Missense | Lefèvre et al. (2006) | ||
| Lugassy (2008), family 2 | 14 | c.1563G > A | p.Trp521* | homo | Nonsense | Lugassy et al. (2008) | |
| Israeli (2013), family 16 | 6 | c.429dup | p.Leu144Alafs*7 | het | Nonsense | Israeli et al. (2013) | |
| 6 | c.466C > T | p.Arg156Cys | het | Missense | Israeli et al. (2013) | ||
| Feng (2017), family 1 | 8 | c.844C > T | p.Arg282Trp | homo | Missense | Bučková et al. (2016) | |
| Feng (2017), family 2 | 6 | c.466C > T | p.Arg156Cys | homo | Missense | Israeli et al. (2013) | |
| Noguera-Morel (2016), family 1 | 8 | c.728G > A | p.Arg243His | homo | Missense | Lefèvre et al. (2006) | |
| Noguera-Morel (2016), family 2 | 12 | c.1303C > T | p.His435Tyr | homo | Missense | Lefèvre et al. (2006) | |
| Scott (2013), patient 1 | 8 | c.712G > A | p.Ala238Thr | homo | Missense | Scott et al. (2013) | |
| Diociaiuti (2016), ID-14 | 8 | c.728G > A | p.Arg243His | het | Missense | Lefèvre et al. (2006) | |
| 10 | c.1084C > T | p.Arg362* | het | Nonsense | Diociauti et al. (2006) | ||
| Diociaiuti (2016), ID-15 | 4 | c.367+1G > A | p.spl? | het | Splice | Diociauti et al. (2006) | |
| 12 | c.1303C > T | p.His435Tyr | het | Missense | Lefèvre et al. (2006) | ||
| Bučková (2016), patient 24 | 3 | c.59dup | p.Ile21Hisfs*59 | homo | Nonsense | Bučková et al. (2016) | |
| Bučková (2016), patient 25 | 3 | c.59dup | p.Ile21Hisfs*59 | homo | Nonsense | Bučková et al. (2016) | |
| Bučková (2016), patient 26 | 8 | c.844C > T | p.Arg282Trp | het | Missense | Bučková et al. (2016) | |
| 10 | c.1085G > A | p.Arg362Gln | het | Missense | Bučková et al. (2016) | ||
| Bučková (2016), patient 27 | 10 | c.1085G > A | p.Arg362Gln | homo | Missense | Bučková et al. (2016) | |
| Palamar (2015), patient 1 | 9 | c.976C > T | p.Arg326* | homo | Nonsense | Palamar et al. (2015) | |
| Gruber (2017), family 1 | 6 | c.549+5G > C | p.spl? | homo | Splice | Gruber et al. (2017) | |
| Sugiura (2013), patient 1 | 11 | c.1138del | p.Asp380Thrfs*3 | het | Nonsense | Sugiura et al. (2013) | |
| 8 | c.728G > A | p.Arg243His | het | Missense | Lefèvre et al. (2006) |
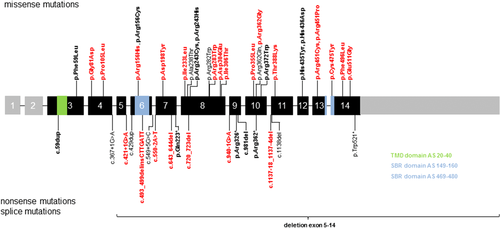
We have shown that there are specific amino acid positions where different pathogenic amino acid changes and recurrent mutations can occur. It should be noted that there is no evidence for a larger hotspot region or domain, since the mutations are evenly distributed throughout the gene. It is noteworthy, that in the putative trans-membrane domain (TMD, amino acids 20–40) and substrate-binding regions (SBR, amino acids 149–160 and 469–480), which should have critical functions for the protein, mutations have not been found more frequently than in other regions. However, a larger cohort is necessary to make significant statistical statements.
4 GENOTYPE–PHENOTYPE CORRELATION
To date, information about genotype–phenotype correlations for ARCI with CYP4F22 mutations is limited in the literature. In general, patients with CYP4F22 mutations often present a more erythrodermic status of the skin at birth and a milder LI or CIE phenotype later in life. It is not clear, why some patients with CYP4F22 mutations show a milder phenotype, whereas others show more severe features of ARCI, in particular when they are born as a collodion baby. Mild and diffuse palmoplantar keratoderma with pronounced lichenification can be found in some patients, whereas others only show hyperlinearity. Especially noteworthy are several cases of SICI in the literature (e.g., Diociaiuti et al. 2016, Noguera-Morel et al., 2016, Sugiura et al., 2013, Virolainen et al., 2000) and in our cohort. We have access to detailed clinical data about the skin status at birth from 38 patients of our cohort. Nineteen of them were born as CB, and 19 patients presented other ichthyotic skin manifestations at birth. In the literature, patients with CYP4F22 mutations who were born with a collodion membrane have been repeatedly reported (Diociaiuti et al. 2016, Feng et al., 2017, Gruber et al. 2017, Lugassy et al., 2008, Noguera-Morel et al., 2016, Sugiura et al., 2013). The phenotype in patients with CYP4F22 mutations is relatively mild, however, the occurrence of a collodion membrane seems to be a common feature. A selection of photos from our patients is shown in Figure 2.
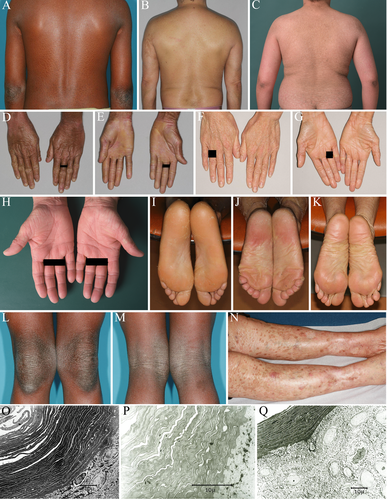
We examined whether the type of the mutation is correlated with the phenotype. In other ARCI genes, nonsense, frameshift or splice-site mutations can lead to more severe phenotypes, e.g., collodion membranes at birth or more severe affected adults. We studied our patients and patients from the literature but did not find any indication for an association between the type of the mutation and the severity of the phenotype. A remarkable example was reported by Gruber et al. (2017). They presented two siblings with ARCI, both carrying the same homozygous splice-site mutation in CYP4F22. One girl was born as CB with contractures of the great joints due to tightened skin, whereas the sister was born with dry skin without a collodion membrane. At the age of 6 months, both sisters showed a generalized fine-scaling phenotype. However, a residual amount of a functional transcript in one case could not be excluded by the authors. In addition, several patients of our study and in the literature presented collodion membranes carrying missense mutations. Besides nonsense, frameshift or splice-site mutations also missense mutations in CYP4F22 have been described in patients that were born as CB both in our study as well as in literature. Consequently, there is no evidence to date that the type of mutation in CYP4F22 does affect the ichthyosis phenotype.
We examined patients with missense mutations affecting the putative trans-membrane domain and substrate-binding regions to consider whether these mutations may lead to a more severe phenotype, since these regions are important for the function of the protein. Feng et al. (2017) and Sugiura et al. (2013) assumed that mutations affecting SBRs may be associated with the development of collodion membranes at birth. In the SBR domain, the known missense mutation p.Arg156Cys and the novel missense mutations p.Arg156His and p.Cys475Tyr are located (Figure 1). In the TMD domain, the duplication c.59dup is located, but a missense mutation has not been identified so far. P49 is compound heterozygous for the mutations p.Arg156Cys and p.Arg156His, for this patient no clinical data were available. P53 is compound heterozygous for p.Cys475Tyr and p.Asp304Glu and was born as a CB. At the present time, there is no clear evidence for a more severe phenotype when one or both mutations are located in these domains.
Interestingly, two patients (P10 and P11) developed skin carcinomas (communicated by E.B.). Both patients had adult congenital ichthyosiform erythroderma, unusual erythematous papules on the legs and arms and phototype II and III. P11 developed several kerathoacanthomas (more than 40 lesions, 2–3 surgeries per year), pre-epitheliomatous keratosis and well-differentiated squamous cell carcinomas on the legs from the age of 43. P10 had one nodular basal cell carcinoma on the internal canthus of the left eye and two pre-epitheliomatous keratosis on the left arm and behind the left ear from the age of 45. It is not clear, whether erythematous papules could be a predictor of skin cancer in patients with CYP4F22 mutations, especially since biopsy of papular and not papular ichthyosis skin showed identical histological features (P10 and P11). In the other patients of our cohort, no observations of skin cancer were mentioned, but the occurrence of numerous cutaneous epidermoid carcinomas has already been reported during hereditary ichthyosis; the cases reported in the literature were not genetically characterized (Natsuga, Akiyama, & Shimizu, 2011).
5 ELECTRON MICROSCOPY
We performed electron microscopy analysis in the skin of P52 and P53 (Figure 2). All three biopsies looked alike, presenting acanthosis and orthohyperkeratosis, normal granular layer, numerous corneodesmosomes, also between outer corneocytes, but no specific inclusions within the horny lamellae nor specific ultrastructural aberrations of structural proteins or organelles involved in terminal differentiation/keratinization (as in the case, e.g., for mutations in NIPAL4, ABCA12, TGM1, FATP4, PNPLA1, e.g. Oji et al., 2010). Increased lamellar body numbers as described by Gruber et al. (2017) seemed not to be a consistent feature, but can be observed in several keratinization disorders (personal communication by I.H.).
6 PERSPECTIVES AND CONCLUSIONS
In summary, we expand the known clinical and molecular spectrum of patients with ARCI due to CYP4F22 mutations. The update reveals recurrent mutations. Moreover, 23 previously unreported pathogenic mutations in CYP4F22 are described.
Mutations in CYP4F22 were detected in about 10% in our cohort of 770 families. Patients with CYP4F22 generally present a mild to moderate ARCI phenotype. As for other subtypes of ARCI, the phenotypic spectrum is broad. We suspect that there may be genetic, epigenetic, and environmental modifiers. Hence, it is recommended to analyze ARCI patients with NGS via multigene panels, which include all known ARCI genes.
Interestingly, patients with CYP4F22 mutations may have a higher risk of developing skin malignancies. Especially in patients with multiple erythematous papules, a regular cancer screening is recommended. However, a direct association between CYP4F22 mutations and skin malignancies cannot be assumed currently and requires further studies.
ACKNOWLEDGMENTS
The authors are grateful to the patients and their families for supporting this study. We would like to thank Susan Cure for critical comments on the manuscript.
CONFLICTS OF INTEREST
The authors declare that there is no conflict of interest.