Next generation sequencing identifies double homozygous mutations in two distinct genes (EXPH5 and COL17A1) in a patient with concomitant simplex and junctional epidermolysis bullosa
Funding information:
Grant Support: debra International (Uitto2)
Communicated by Peter H. Byers
Abstract
Epidermolysis bullosa (EB) is a heterogeneous group of heritable blistering diseases. We developed a next generation sequencing (NGS) panel covering 21 genes associated with skin fragility disorders, and it was applied to DNA from 91 probands with the diagnosis of EB. In one patient, novel homozygous mutations were disclosed in two different, unlinked EB-associated genes: EXPH5, chr11 g.108510085G > A; p.Arg1808Ter and COL17A1, chr10 g.104077423delT; p.Thr68LeufsTer106. Consequences of the COL17A1 mutation were examined by RNAseq which revealed a complex splicing pattern predicting synthesis of a truncated polypeptide (85%) or in-frame deletion of exon 4 (15% of transcripts). Transmission electron microscopy (TEM) and immunostaining revealed findings consistent with EB simplex (EBS) and junctional EB (JEB), and clinical examination revealed a complex phenotype with features of both subtypes. This case illustrates the power of next generation sequencing in identifying mutations in patients with complex EB phenotype, with implications for genotype–phenotype correlations, prenatal testing, and genetic counseling of families at risk for recurrence.
Next generation sequencing (NGS) applications have entered into the clinical diagnostic setting, and whole exome sequencing (WES) now offers an important tool for implementation of precision medicine. The diagnostic WES provides the opportunity to investigate the relationships between multi-locus genomic variations and complex phenotypes, such as the presence of more than one Mendelian disease in one patient. Previous reports have shown the existence of multiple genetic diseases in up to 5% of the genetic patients referred to studies (Monies et al., 2017; Posey et al., 2017; Trujillano et al., 2017). In most cases, two diseases in a person have been recognized with distinct patterns of clinical manifestations, but in some occasions, concurrent diseases have overlapping phenotypic features. Within this spectrum, there is also a possibility of co-existence of two mutated genes belonging to a disease in case of conditions with genetic heterogeneity, such as the case with epidermolysis bullosa (EB) presented here.
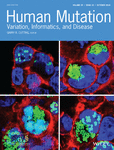
The heritable forms of EB manifest at birth or shortly thereafter with blistering and erosions of the skin and mucous membranes, with considerable morbidity and mortality (Fine et al., 2014). On the basis of clinical observations and ultrastructural demonstration of the topographic level of blistering within the skin, EB has been divided into four broad categories: (a) EB simplex (EBS) demonstrates intra-epidermal blistering; (b) junctional EB (JEB) depicts tissue separation within the dermal–epidermal basement membrane; (c) the dystrophic forms (DEB) demonstrate sub-lamina densa blister formation within the upper papillary dermis; (d) and Kindler syndrome which can demonstrate multiple levels of tissue separation even in the same individual. There is extensive heterogeneity in the severity of the phenotype and the outcome of the disease in these subgroups of EB. The phenotypic heterogeneity of different forms of EB reflects the fact that as many as 20 different genes encoding the components of the dermal–epidermal attachment complexes harbor mutations in different subtypes of EB (Uitto et al., 2017). Recently, a number of NGS approaches have been adopted, including gene-targeted panels, as well as whole exome sequencing and whole genome sequencing (WGS) for study of EB (Takeichi et al., 2015; Vahidnezhad, Youssefian, Saeidian, et al., 2017; Vahidnezhad, Youssefian, Zeinali, et al., 2017). We have developed a NGS panel which consists of 21 genes associated with different skin fragility syndromes, including 19 genes shown to harbor mutations in patients with EB. Sequencing of a large number of EB patients, primarily from consanguineous marriages in Iran, identified a number of novel, previously unreported mutations in a number of cases with unusual genetic constellations and complex cutaneous phenotypes not diagnostic of any of the particular subtype of EB (Vahidnezhad, Youssefian, Saeidian, et al., 2017). In this study, we report identification of homozygous mutations in two distinct genes associated with different subtypes of EB resulting in an unusual cutaneous phenotype (Figure 1a).
This study was approved by the Institutional Review Board of the Pasteur Institute of Iran, and the individuals, as well as parents of underage patients, gave a written informed consent to participate in the research and permission to publish their images. For targeted next-generation sequencing with a 21-gene array, DNA was isolated from peripheral blood samples taken from EB patients, their parents and other clinically affected and unaffected family members. For details of the library, data capturing and bioinformatics, see Supplementary Material and Methods.
For RNAseq, total RNA was extracted from a skin biopsy, and sequencing reads were generated on the Illumina NextSeq 500 platform and stored in FASTQ format. For downstream data analysis, including mapping of regions of homozygosity (ROH), see Supplementary Material and Methods.
Transmission electron microscopy (TEM) and immunofluorescence (IF) staining were performed as described previously (Vahidnezhad et al., 2018). For the antibodies used in this study, see Supp. Table S1.
The NGS array developed by us covers 21 genes associated with skin fragility disorders (Vahidnezhad, Youssefian, Saeidian, et al., 2017). The sequencing panel was applied to DNA from 91 probands with the clinical diagnosis of EB, based on neonatal blistering and erosions of the skin. In one patient, two potentially pathogenic sequence variants were disclosed: (a) a homozygous GRCh38 chr11 g.108510085G > A (c.5422C > T; p.R1808Ter) mutation was found in exon 6 (Refseq: NM_015065.2) of the EXPH5 gene; and (b) a homozygous sequence variant c.202delA; p.T68LfsTer106 was discovered in exon 4 (RefSeq: NM_000494.3) of the COL17A1 gene (Figure 1). These novel mutations have been submitted to LOVD v.3.0-Leiden Open Variation Database (https://databases.lovd.nl/shared/variants/0000165326). The parents were heterozygous for the respective sequence variants. These previously unreported mutations were confirmed by bidirectional Sanger sequencing (Figure 1d), and they are not present in ExAC or 1000 Genomes databases. Therefore, the chance of coinheritance of these mutations in homozygous form is extremely low. The deleted A in the c.202delA allele is the last nucleotide of exon 4 at the exon 4/intron 4 border and results in replacement of the donor splice junction sequence AA/gt in the control with CA/gt in the patient (Figure 1d). This mutation was predicted to result in frame-shift and/or aberrant splicing. The mutations in COL17A1 and EXPH5 were shown by genome-wide homozygosity mapping to reside within regions of homozygosity of 20.4 and 17.4 Mb in chromosomes 10 and 11, respectively (Figure 1c). It should be noted that in the cohort of 91 patients, no other patients with biallelic homozygous mutations in two different genes were identified. Previously, a family with mutations in COL17A1 and LAMB3 have been described (Floeth & Bruckner-Tuderman, 1999); mutations in these two genes are associated with the junctional form of EB and cause altered expression of the components of the hemidesmosome-anchoring filament complex. In our case, exophilin 5 and type XVII collagen, encoded by EXPH5 and COL17A1, reside in different compartments of the dermal–epidermal junction, but their deficiency, either alone or in combination, could explain reduction of keratin 14 and plectin, as demonstrated by immunostaining.
To examine the consequences of the c.202delA mutation on splicing, the COL17A1 mRNA expression profile was examined by RNAseq using total RNA isolated from a skin biopsy from the patient and a matched control. Sashimi blot of the control RNA revealed canonical splicing out of introns 3 and 4, with no evidence of alternative splicing, indicating 100% retention of exon 4 (Figure 1e). In the patient's mRNA, a complex splicing pattern emerged: The majority of transcripts (85%) extending from the donor site of exon 3 utilized the acceptor site in exon 4, and therefore resulted in a transcript which was out-of-frame due to deletion of the last nucleotide in exon 4, predicting synthesis of a nonfunctional polypeptide truncated at codon 106 downstream from the mutation. A fraction of the mRNA (∼15%) skipped introns 3 and 4, together with exon 4 (Figure 1e). Since the wild-type exon 4 consists of 105 nucleotides and is, therefore, in frame, this splicing event predicted synthesis of a polypeptide shortened by 35 amino acids due to internal deletion of exon 4. It was also noted that some intron 5 sequences were retained, and that a small number of transcripts skipped exon 6, together with introns 5 and 6.
To examine the consequences of the two mutations at tissue level, skin biopsies obtained from the proband were subjected to histopathology, transmission electron microscopy and immunofluorescence staining. Light microscopy of semi-thin sections revealed cellular disruption within the basal keratinocytes, which appeared pale with loss of keratin filaments and showed cytolysis (Figure 2a). TEM identified blister formation within the lower pole of basal keratinocytes as well as focally within the lamina lucida, consistent with EBS and JEB, respectively (Figure 2b and c). The cytoplasm of some basal keratinocytes contained numerous membrane-bound vesicles, and while the intercellular desmosomes at cell–cell junctions appeared normal, the adjacent keratin displayed compacted filaments as well as intercellular vesicles consistent with EBS (Figure 2d and e). At the dermal–epidermal junction, there was a reduced number of hypoplastic hemidesmosomes consistent with JEB (Figure 2f).
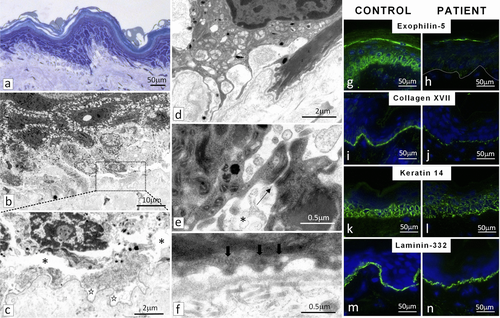
Immunofluorescence staining with an anti-exophilin-5 antibody of the patient's skin was essentially negative (Figure 2g and h). Staining of collagen XVII in the patient's skin was significantly attenuated, with an interrupted pattern (Figure 2i and j). The presence of residual collagen XVII staining in the patient's skin may reflect the synthesis of internally truncated polypeptide devoid of exon 4 sequences, as predicted by the RNAseq results. Keratin 14 labelling in the patient's skin showed an irregular pattern within the epidermis (Figure 2k and l). Immunostaining for laminin-332 demonstrated bright linear labelling at the dermal–epidermal junction both in control and the patient's skin (Figure 2m and n). In addition, staining for collagens IV and VII revealed similar basement membrane labeling both in the patient's and the control skin, while staining with two separate antibodies for plectin, recognizing the amino- and carboxy-terminus, respectively, revealed markedly attenuated, irregular pattern (Supp. Figure S1).
The proband, a 3-year old male, the only child of consanguineous parents, was diagnosed at birth with EB. There was no family history of similar conditions, and specifically, the parents were clinically unaffected. The patient presented with complex EB phenotype with features of both EBS and JEB. For details of the clinical manifestations, see Supplementary Material. Furthermore, TEM revealed tissue separation both within the basal keratinocytes in the epidermis as well as within the lamina lucida of the epidermal basement membrane, the ultrastructural diagnostic features of EBS and JEB, respectively. These observations can be explained by the presence of double homozygous mutations in two distinct genes, EXPH5 and COL17A1, which have been previously shown to harbor mutations in these two subtypes of EB, respectively. The presence of these two homozygous mutations in two distinct genes in this patient apparently reflects the consanguinity in the family, and these mutations were shown to reside within large ROHs in chromosomes 10 and 11, respectively.
Recent retrospective studies have addressed the issue of the presence of mutations in different genes in patients with complex phenotypes. For example, the study by Posey et al. (2017) reanalyzed the WES data from a series of 7,374 consecutive unrelated patients and found a molecular diagnosis in 2,076 (28.2%); among them, 101 (4.9%) patients had more than one disease. In these patients, combinations of different inheritance patterns were observed, but blended phenotypes among these patients with dual diagnosis were either distinct, affecting different organ systems, or overlapping with similar phenotypic features. The majority of pathogenic variants in autosomal dominant disease genes and half of the pathogenic variants in X-linked disease were de novo variants. Only nine patients had at least two autosomal recessive disorders, and in eight of them there was a history of parental consanguinity (Posey et al., 2017 #4026). A study by Trujillano et al. (2017) identified pathogenic or likely pathogenic sequence variants in two different genes in three patients out of 307 families (∼1%). Similarly, Monies et al. (2017) reported that dual molecular diagnosis was rare and only accounted for 1.5% of cases subjected to WES. These relatively low numbers could be attributed to the methodology because dual diagnosis was considered only if both molecular lesions could be classified as at least likely pathogenic.
We have previously reported the co-existence of phenylketonuria either with a maple syrup urine disease or Sandhoff disease in two patients in Iran (Abiri et al., 2016). Quite recently, homozygous mutations in RSPO4 and PADI3 associated with congenital anonychia and uncombable hair syndrome have been reported in a Kuwaiti family (Hsu et al., 2017). In these cases, no patients with homozygous pathogenic mutations in two genes related to one disease, as in our patient with EXPH5 and COL17A1 mutations associated with EB, were noted. Previous reports have suggested digenic inheritance with mutations in the KRT5 and KRT14 genes in families with EBS (Kim, Harris, Hyland, & Murrell, 2017; Padalon-Brauch et al., 2012; Wertheim-Tysarowska et al., 2014). However, in two of these studies one of the parents, a heterozygous carrier, showed similar blistering phenotype as the proband, consistent with the fact that the majority of the keratin mutations are dominant and that semi-dominance in a large family with EBS has been reported (Vahidnezhad et al., 2016). In our family, the proband had two homozygous mutations in two distinct, genetically unlinked genes, and the parents, heterozygous carriers of the corresponding genes, were clinically normal, attesting to the recessive nature of the mutations and excluding digenic inheritance.
The practical implication of our studies is that they challenge the practice in which the diagnostic evaluation is halted after identification of the first pathogenic mutations. This has been particularly true when candidate gene approach has been taken to interrogate individual genes by traditional Sanger sequencing approaches. Evidence for mutations causing more than one Mendelian disease in a patient may be derived from deep phenotyping of the cases (Pena et al., 2018) but our report here highlights the possibility of existence of more than one mutated gene within the spectrum of a single disease. Specifically, in our case, both mutations were homozygous and were predicted to result in the absence (EXPH5) or very low levels (COL17A1) of the corresponding protein products, with ultrastructural findings in the skin consistent with the presence of two subtypes, the simplex and the junctional forms, of the disease. Possibly, this phenomenon may be more prevalent than currently recognized, and there may be pathogenic variants which currently have been classified as variants of unknown significance, which may contribute to the pathogenesis of the disease. Finally, the knowledge of both pathogenic mutations in our patient has implications for attempts to establish phenotype–genotype correlations and can impact prenatal testing and genetic counseling of families at risk for recurrence.
ACKNOWLEDGMENTS
The authors would like to thank Drs. Adam Ertel and Paolo Fortina for assistance in next generation sequencing. Carol Kelly assisted in manuscript preparation. This study is in partial fulfillment of the PhD Thesis of HV.
DISCLOSURE STATEMENT
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
HV and LY performed NGS and RNA sequencing, data analysis, experimental design and project planning, and drafting of the manuscript. AG, PAL, LL, and JM performed tissue staining, electron microscopy, and also contributed to data analysis and review of literature. AHS and AT contributed to sequencing, analysis, and figure generation. SS contributed to sample collection, clinical diagnosis, and project planning. SZ and AK provided genetic counseling and experimental contributions. AJ performed bioinformatics analyses. JU oversaw the project and contributed to overall coordination of the project and provided clinical expertise regarding the described case. All authors read and approved the final manuscript.