Missense mutations have unexpected consequences: The McArdle disease paradigm
Communicated by Hans R. Waterham
Abstract
McArdle disease is a disorder of muscle glycogen metabolism caused by mutations in the PYGM gene, encoding for the muscle-specific isoform of glycogen phosphorylase (M-GP). The activity of this enzyme is completely lost in patients’ muscle biopsies, when measured with a standard biochemical test which, does not allow to determine M-GP protein levels. We aimed to determine M-GP protein levels in the muscle of McArdle patients, by studying biopsies of 40 patients harboring a broad spectrum of PYGM mutations and 22 controls. Lack of M-GP protein was found in muscle in the vast majority (95%) of patients, irrespective of the PYGM genotype, including those carrying missense mutations, with few exceptions. M-GP protein biosynthesis is not being produced by PYGM mutations inducing premature termination codons (PTC), neither by most PYGM missense mutations. These findings explain the lack of PYGM genotype–phenotype correlation and have important implications for the design of molecular-based therapeutic approaches.
1 INTRODUCTION
McArdle disease (glycogen storage disease type V; MIM# 232600) is an autosomal recessive disorder caused by pathogenic mutations in the PYGM gene, encoding the muscle-specific isoform of glycogen phosphorylase (M-GP, also known as “myophosphorylase”), which catalyzes the first step of muscle glycogen breakdown. The disease usually provokes exercise intolerance, in the form of acute ‘‘crises’’ of early fatigue, myalgia, and contractures that can be accompanied by rhabdomyolysis and myoglobinuria (Nogales-Gadea et al., 2015; Santalla et al., 2017). Although ∼150 pathogenic PYGM mutations have been described (Nogales-Gadea et al., 2015), no genotype–phenotype correlation has so far been identified in independent cohorts (Andrea Martinuzzi et al., 2003; Quinlivan et al., 2010; Santalla et al., 2017). In the last update of Spanish McArdle patients (n = 333), we reported negative histochemical staining for M-GP activity in 100% of the studied biopsy samples (n = 205 patients) despite considerable heterogeneity in PYGM genotype (Santalla et al., 2017).
Nearly 35% of all reported PYGM mutations result in premature termination codons (PTC), of which the commonest pathogenic variant in Caucasians is the c.148C > T (p.R50*) mutation. A cellular surveillance mechanism, “nonsense-mediated mRNA decay” (NMD) degrades most PTC-containing PYGM transcripts, preventing the synthesis of M-GP (Nogales-Gadea et al., 2008). Nevertheless, ∼50% of reported PYGM mutations are non-synonymous (i.e., missense) (Nogales-Gadea et al., 2015) and yet their functional consequences do not differ from that of PTC, insofar as they do not result in a milder phenotype, as would be theoretically expected (Santalla et al., 2017). Missense PYGM mutations have a widespread distribution across all the domains of the M-GP protein, including glycogen storage site, catalytic site, and dimerization domains (Nogales-Gadea et al., 2015), suggesting that every domain is critical for enzyme activity.
No study has assessed the effect of missense mutations on the translation of M-GP. Previous reports showing M-GP protein expression data were done at a time when McArdle's diagnosis was based on muscle biopsy, as PYGM genotyping was not available (Martinuzzi et al., 1993; Servidei et al., 1988). Here, we aimed to corroborate that absence of M-GP protein in muscle is a hallmark of this disorder irrespective of PYGM genotype.
2 SUBJECTS AND METHODS
The study was approved by the local ethics committee and was performed in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants. Muscle biopsies were obtained from 40 patients and 22 controls with normal histochemical muscle biopsy results. All patients were diagnosed with McArdle disease (i.e., homozygous/compound heterozygous for documented pathogenic PYGM mutations, except for patient P23, in whom only one mutant allele was identified but who presents all the signs and symptoms of the disease, including the “second wind” phenomenon (Table 1) (Vissing & Haller, 2003). Muscle biopsy results showed both negative staining in the histochemical reaction for M-GP and null activity for the enzyme (data available from 16 patients), except for P10, whose muscle biopsy was taken shortly after an exertional rhabdomyolysis episode – such episodes induce re-expression of non-muscle GP isoforms, which may lead to “false-positive” results for M-GP expression (Nogales-Gadea et al., 2015)
| Patient | PYGM biallelic genotype | Age (y) | Sex | M-GP staining | GP activity (μmol/min/g muscle tissue)* | PYGM RNA expression (% of controls) | DIGE-MALDI TOF/TOF M-GP peptides | LC-MS/MS M-GP peptides |
|---|---|---|---|---|---|---|---|---|
| 1 | c.2310_2311dupCC/ c.2310_2311dupCC | 52 | M | Negative | 0 | 70% | ||
| 2 | c.2392T > C/c.2392T > C | 55 | F | Negative | 0 | 77% | ||
| 3 | c.148C > T/c.148C > T | 35 | F | Negative | 0 | 1%(4) | ||
| 4 | c.148C > T/c.148C > T | 21 | M | Negative | 0 | NR | ||
| 5 | c.148C > T/c.645G > A | 28 | M | Negative | 0 | 5%(17) | ||
| 6 | c.148C > T/c.2392T > C | 14 | M | Negative | 0 | 70%(4) | ||
| 7 | c.2392T > C/c.2392T > C | 56 | M | Negative | 0 | 55%(4) | ||
| 8 | c.148C > T/c.280C > T | 34 | M | Negative | 0 | 41%(4) | ||
| 9 | c.613G > A/c.613G > A | 39 | M | Negative | NR | NR | ||
| 10 | c.148C > T/c.1094C > T | 0 | M | Negative | 5.5 | NR | ||
| 11 | c.613G > A/c.613G > A | 32 | M | Negative | 0 | NR | ||
| 12 | c.521G > A/c.1827 G > A | 25 | M | Negative | 0 | 59% | ||
| 13 | c.148C > T/c.13_14delCT | 28 | F | Negative | 0 | 4%(4) | ||
| 14 | c.2392T > C/c.2392T > C | 45 | M | Negative | 0 | NR | ||
| 15 | c.148C > T/c.148C > T | 63 | M | Negative | 0 | NR | ||
| 16 | c.148C > T/c.1979C > A | 29 | M | Negative | 0 | 45%(4) | ||
| 17 | c.2392T > C/c.2392T > C | 30 | F | Negative | NR | NR | ||
| 18 | c.148C > T/c.148C > T | 19 | F | Negative | NR | NR | ||
| 19 | c.148C > T/c.1147G > A | 75 | M | Negative | NR | NR | ||
| 20 | c.613G > A/c.613G > A | 14 | M | Negative | NR | NR | Absent | NR |
| 21 | c.148C > T/c.2392T > C | 61 | M | Negative | NR | NR | Absent | NR |
| 22 | c.148C > T/c.148C > T | 71 | M | Negative | NR | NR | Absent | Absence |
| 23 | c.1470dupG/? | 41 | M | Negative | NR | NR | ||
| 24 | c.148C > T/c.2262delA | 34 | M | Negative | NR | NR | ||
| 25 | c.13_14delCT/c.13_14delCT | 59 | F | Negative | NR | NR | ||
| 26 | c.148C > T/c.148C > T | 54 | M | Negative | NR | NR | ||
| 27 | c.613G > A/c.613G > A | 42 | M | Negative | NR | NR | ||
| 28 | c.2392T > C/c.2392T > C | 13 | F | Negative | NR | NR | ||
| 29 | c.13_14delCT/c.13_14delCT | 52 | F | Negative | NR | NR | ||
| 30 | c.1092-1G > T/c.244-3_244-2CA | 57 | M | Negative | NR | NR | ||
| 31 | c.148C > T/c.148C > T | 59 | F | Negative | NR | NR | Absent | Absent |
| 32 | c.2262delA/c.2262delA | 32 | F | Negative | NR | NR | Absent | Absent |
| 33 | c.148C > T/c.1366G > A | 34 | F | Negative | NR | NR | Present | Present |
| 34 | c.148C > T/c.613G > A | 52 | F | Negative | NR | NR | NR | Absent |
| 35 | c.2392T > C/c.2392T > C | 48 | M | Negative | NR | NR | NR | Absent |
| 36 | c.148C > T/c.2111C > T | 55 | M | Negative | NR | NR | NR | Absent |
| 37 | c.148C > T/c.347T > C | 45 | M | Negative | NR | NR | NR | Absent |
| 38 | c.13_14delCT/c.2262delA | 29 | F | Negative | 0 | 5% | ||
| 39 | c.613G > A/c.613G > A | 60 | M | Negative | 0 | 148% | ||
| 40 | c.2310_2311dupCC/c.2310_2311dupCC | 55 | M | Negative | NR | 77% |
- Results of analysis of peptides related to M-GP by two mass spectrometry methods, matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF/TOF) and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) are also shown.
- Missense mutations are in bold and results for these variants are shown in grey background. Abbreviations: F, female; M, male; NR, not reported.
- Symbols: ?, not found with Sanger sequencing of PYGM gene intron/exon boundaries. *Controls’ muscle glycogen phosphorylase activity (n, mean±SD) (35, 23.5±4.0).
Muscle biopsies were subjected to immunoblotting using three different primary antibodies against M-GP: a goat (Martinuzzi et al., 1993) and two rabbit polyclonal antibodies (ab63158; Abcam, Cambridge, UK; and HPA056003; Sigma-Aldrich, St. Louis, MO). As a control, membranes were probed with an antibody against the housekeeping protein α-tubulin (T6074; Sigma-Aldrich). Membranes were developed with ECL (Amersham, Buckinghamshire, UK). Images were obtained with the LAS-3000 system (FujiFilm, Tokyo, Japan) and quantified with NIH ImageJ (v.1.37) software (Scion image, NIH). The Mann–Whitney U-test was used for between-group comparisons (α = 0.05).
For proteomic analysis, frozen muscle tissue samples (15 mg) were manually homogenized in an ice water bath in 1/10 1 × TBS–1% SDS buffer including protease inhibitors, and then centrifuged (1 min, 14,000 rpm). Protein quantification of supernatants was determined using the MicroBCA Protein Assay Kit (ThermoFisher, Waltham, MA). Analysis was performed with high-performance liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) (Moreno et al., 2014; Shevchenko, Wilm, Vorm, & Mann, 1996). Searches against the decoy database were done using a false discovery rate <0.01 (Alonso et al., 2015). We also performed two-dimensional difference gel electrophoresis (2D-DIGE) as previously reported (Martin-Lorenzo et al., 2014) in six controls and six patients. Differential proteins were tryptic-digested and identified in a 4800 plus MALDI-TOF/TOF (Proteomics Analyzer) mass spectrometer with 4000 Series Explorer™ v.3.5 software (Applied Biosystems/MDS Sciex, Toronto, Canada) (Sechi & Chait, 1998), with a probability score greater than that fixed by Mascot as being significant (p < 0.05).
Polysomal profiling and RNA analysis was performed as detailed elsewhere (Martinez-Azorin, Remacha, & Ballesta, 2008). Myoblasts were isolated from biopsies of P40 and a control subject by explant culture (Askanas & Gallez-Hawkins, 1985). Retrotranscription was done using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA), and amplification of the cDNA corresponding to the PYGM gene (NM_005609.2) was performed in two fragments as previously reported (Garcia-Consuegra et al., 2016). One microliter of amplified PYGM cDNA encompassing the position of the mutation c.2310_2311dupCC (p.R771Pfs*33) was analyzed on the Agilent 2100 Bioanalyzer in combination with microfluidic DNA-1000 chips (Agilent Technologies, Santa Clara, CA).
3 RESULTS
The c.148C > T (p.R50*) variant was the most prevalent PYGM pathogenic mutation in our cohort (Table 1). Missense mutations were also prevalent (55% of patients), and included the two other more frequent PYGM mutations in the Spanish population, c.2392T > C (p.W798R) and c.613G > A (p.G205S), and the non-synonymous variants c.280C > T (p.R94W), c.347T > C (p.L116P), c.521G > A (p.G174D), c.1094C > T (p.A365V), c.1147G > A (p.E383K), c.1366G > A (p.V456M), c.1979C > A (p.A660D), and c.2111C > T (p.A704V). Four frameshift PTC c.13_14delCT (p.L5Vfs*22), c.2262delA (p.K754Nfs*49), c.2310_2311dupCC (p.R771Pfs*33), c.1470dupG (p.R491AfsX7) and four splice-disrupting mutations c.645G > A, c.1827 G > A, c.1093-1G > T (Bruno et al., 2006) and c.244-3_244-2CA) (Fernandez-Cadenas et al., 2003; Garcia-Consuegra et al., 2016) were also identified. All these variants have been deposited in the Leiden Open Variation Database, and can be found in the following link: https://databases.lovd.nl/shared/variants/PYGM?search_var_status=%3D%22Marked%22%7C%3D%22Public%22. Data on PYGM RNA expression was available from 13 patients. The NMD pathway was presumably acting on all the PTC-generating mutations, except in P40 (c.2310_2311dupCC homozygous, displaying 77% of normal transcript levels – a finding not unexpected as this mutation should bypass NMD control owing to its location (last PYGM exon)). For missense mutations, gene expression levels were consistently ≥40% of control values.
Immunoblots from patients with PTC-predicting mutations (c.13_14delCT, c.2262delA, and c.2310_2311dupCC (Table 1)) showed the total absence of M-GP protein (Figure 1A). Remarkably, this finding also held true for the vast majority of patients harboring missense mutations in heterozygosity/homozygosity, in whom normal protein translation and biosynthesis of M-GP was theoretically expected. These results were consistent irrespective of which of the three antibodies against M-GP was used (Table 1, Figure 1A). Only two patients, both carriers of a missense mutation in trans with c.148C > T, c.1147G > A (P19) and c.1366G > A (P33), had M-GP protein levels similar to those of controls. Overall, mean relative levels of M-GP protein were significantly lower (p < 0.001) in patients (0.087±0.04) than in controls (1.20±0.06) (Figure 1B). Additionally, M-GP assessed by 2D-DIGE corroborated the presence of M-GP in P33 (Figure 1C).
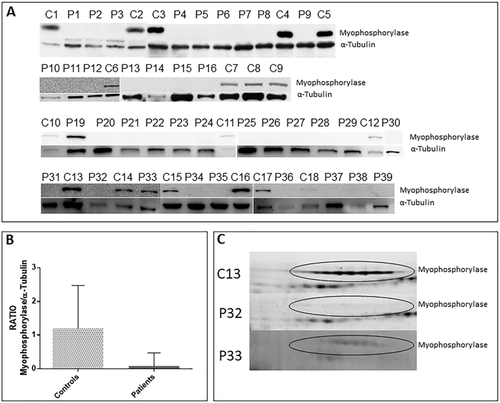
To confirm that the band corresponding to the molecular weight of wild-type M-GP found by immunoblotting in P33 was specifically M-GP, the remaining muscle tissue sample was analyzed by LC-MS/MS and MALDI-TOF/TOF, which showed only specific peptides matching human M-GP (Table 1), excluding the possibility that the protein detected by immunoblotting was a non-muscle (i.e., brain, liver) isoform. No remaining muscle tissue from P19 was available for confirmation analyses. In nine patients who showed the absence of M-GP by immunoblot, no residual peptides of M-GP were found by the two proteomic approaches, supporting the impairment in M-GP expression.
To gain insight into the mechanism by which M-GP expression is impaired by non-NMD-affected PYGM mutations, we studied the polysomal cell fraction in cultured myotubes established from P40, allowing us to determine whether the PYGM transcript is synthesized in polyribosomes (Martinez-Azorin et al., 2008). Patient 40 harbored the homozygous frameshift mutation c.2310_2311dupCC, which would be expected to bypass NMD surveillance (Nogales-Gadea et al., 2010) and M-GP translation should occur normally; however, no M-GP protein was detected by immunoblotting. Both ribosomal fractions belonging to single ribosomes (40S + 60S + 80S) and the polysomal fraction (belonging to multiple ribosomes) bound to the PYGM transcript were isolated (Suppl. Figure S1, upper panel) and PYGM cDNA was analyzed on microfluidic chips. PYGM cDNA was detected in the polysomal fraction to an extent similar to that of the control (compare Suppl. Figure S1 C and D), but an apparent lower intensity was found in the patient PYGM cDNA bound to single ribosomes (compare Suppl. Figure S1 A and B).
4 DISCUSSION
M-GP protein in skeletal muscle tissue is absent in the vast majority of McArdle patients of this cohort (95%) with a broad spectrum of PYGM mutations, including several missense mutations (10 of ∼80 known missense PYGM mutations and affecting different domains of the M-GP protein) (Nogales-Gadea et al., 2015). It thus seems likely that suppression of M-GP expression is a commonly elicited mechanism in the muscle tissue of McArdle patients. Missense mutations could likely alter PYGM mRNA secondary structure, leading to ribosome stalling and subsequent degradation, as suggested from our findings in cultured myoblasts from a patient carrying a homozygous frameshift mutation escaping NMD and having an apparent decrease of PYGM transcripts bound to single ribosomes.
Our results suggest that the documented lack of correlation between PYGM genotype and individual phenotype is due to the absence of M-GP enzyme activity resulting from impaired biosynthesis, not only for PTC-containing mutations, but also for most missense mutations. These findings add to the current knowledge of the molecular mechanisms underlying this and potentially other muscle genetic diseases and may have important implications for the design of molecular-based therapeutic approaches. In this regard, PTC read-through compounds (e.g., Ataluren) are therapeutically appealing because many McArdle patients harbor “NMD-affected PTC” mutations (Nogales-Gadea et al., 2015). Although these compounds appear to incorporate specific amino acids at a PTC (Roy et al., 2016), studying to what extent the resulting proteins retain normal functionality is important in light of our findings that most PYGM non-synonymous mutations obstruct M-GP biosynthesis.
ACKNOWLEDGMENTS
The research of Gisela Nogales-Gadea, Alejandro Lucia, Joaquin Arenas, and Miguel A. Martin in the field of McArdle disease is funded by Instituto de Salud Carlos III (grant numbers PI15/01756, PI15/00558, PI12/00914, PI14/00903, and PI15/00431) and co-financed by Fondos FEDER. Gisela Nogales-Gadea is supported by a Miguel Servet research contract (ISCIII CP14/00032 and FEDER) and by a Trampoline Grant #21108 from AMF Telethon. Alfonsina Ballester-Lopez is funded by an FI Agaur fellowship ref. FI_B 01090. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data. We gratefully acknowledge Rocio Garrido and Francisco Martinez-Azorín for their technical support, and all the physicians and pathologists who provided samples from their McArdle patients.
AUTHOR CONTRIBUTIONS
I-GC: conception and design of the study, acquisition and analysis of data or drafting, a significant portion of the manuscript and figures; S-AP: acquisition and analysis of data; A-BL: acquisition and analysis of data; R-FV: acquisition and analysis of data; T-P: drafting a significant portion of the manuscript; G-PM: acquisition and analysis of data; J-CM: acquisition and analysis of data; A-GQ: acquisition and analysis of data; AL-A: drafting a significant portion of the manuscript; J-A: drafting a significant portion of the manuscript; A-L: conception and design of the study, and drafting a significant portion of the manuscript and figures; G-NG: conception and design of the study, acquisition and analysis of data and drafting a significant portion of the manuscript and figures; MA-M: conception and design of the study, acquisition and analysis of data or drafting a significant portion of the manuscript and figures.
CONFLICTS OF INTEREST
J. Arenas, A. Lucia, and M.A. Martín declare grants from the Spanish government granting agency Instituto de Salud Carlos III, Madrid, Spain. G. Nogales-Gadea declares grants from Instituto de Salud Carlos III, Madrid, Spain and AMF Telethon, France. G. Pintos-Morell reports personal fees from Shire, and from BioMarin, outside the submitted work. The remaining co-authors declare no conflicts of interest.