Comprehensive Analysis of Pathogenic Deletion Variants in Fanconi Anemia Genes
Contract grant sponsors: Grant R01 HL120922 from NIH, Grant #UL1 TR000043 from NCATS, NIH CTSA program, and NHGRI Intramural Research Program, NIH; Anderson Cancer Center at the Rockefeller University, Burroughs Wellcome Fund Career Award for Medical Scientists; Fanconi Anemia Research Fund.
Communicated by David E. Goldgar
ABSTRACT
Fanconi anemia (FA) is a rare recessive disease resulting from mutations in one of at least 16 different genes. Mutation types and phenotypic manifestations of FA are highly heterogeneous and influence the clinical management of the disease. We analyzed 202 FA families for large deletions, using high-resolution comparative genome hybridization arrays, single-nucleotide polymorphism arrays, and DNA sequencing. We found pathogenic deletions in 88 FANCA, seven FANCC, two FANCD2, and one FANCB families. We find 35% of FA families carry large deletions, accounting for 18% of all FA pathogenic variants. Cloning and sequencing across the deletion breakpoints revealed that 52 FANCA deletion ends, and one FANCC deletion end extended beyond the gene boundaries, potentially affecting neighboring genes with phenotypic consequences. Seventy-five percent of the FANCA deletions are Alu–Alu mediated, predominantly by AluY elements, and appear to be caused by nonallelic homologous recombination. Individual Alu hotspots were identified. Defining the haplotypes of four FANCA deletions shared by multiple families revealed that three share a common ancestry. Knowing the exact molecular changes that lead to the disease may be critical for a better understanding of the FA phenotype, and to gain insight into the mechanisms driving these pathogenic deletion variants.
Introduction
Fanconi anemia (FA) is a rare autosomal- or X-linked-recessive disorder, characterized by congenital malformations, bone marrow failure, and predisposition to cancer, particularly hematological malignancies and solid tumors of the head and neck. The classic characteristic of FA cells is the increased sensitivity to DNA-interstrand cross-linking agents such as diepoxybutane [Auerbach, 2009]. FA is genetically heterogeneous, resulting from mutations in one of the 16 known FA genes (FA Mutation Database, http://www.rockefeller.edu/fanconi/), including FANCA (MIM #607139), FANCB (MIM #300515), FANCC (MIM #613899), FANCD1 (MIM #605724)/BRCA2 (MIM #600185), FANCD2 (MIM #613984), FANCE (MIM #613976), FANCF (MIM #613897), FANCG (MIM #602956), FANCI (MIM #611360), FANCJ (MIM #609054)/BRIP1 (MIM #605882), FANCL (MIM #608111), FANCM (MIM #609644), FANCN (MIM #610832)/PALB2 (MIM #610355), FANCO (MIM #613390)/RAD51C (MIM #179617), FANCP (MIM #613951)/SLX4 (MIM #613278), and FANCQ (MIM #615272)/ERCC4 (MIM #133520) [Neveling et al., 2009; Bogliolo et al., 2013]. The proteins encoded by these genes participate in pathways that are involved in detection and resolution of DNA-interstrand cross-links, maintenance of hematopoietic stem cells, and prevention of tumorigenesis [Kottemann and Smogorzewska, 2013].
FANCA, FANCC, and FANCG pathogenic variants account for the disease in 60%, 15%, and 10% of FA families, respectively, whereas mutations in the other genes occur less frequently (0.1%–4%) [Neveling et al., 2009]. The mutation spectrum includes single-nucleotide variations (SNVs), small insertions and deletions (INDELs), and large deletions. Large deletions contribute 20%–40% of all FANCA mutations [Centra et al., 1998; Levran et al., 1998; Morgan et al., 1999; Moghrabi et al., 2009; Castella et al., 2011]. The majority of these deletions were identified by multiplex ligation probe analysis (MLPA), identifying deleted exons only, but not the precise breakpoints.
Although the majority of pathogenic variants associated with FA are private germline variants, a few common founder mutations have been reported [Whitney et al., 1993; Tipping et al., 2001; Auerbach et al., 2003; Callen et al., 2005; Morgan et al., 2005; Castella et al., 2011; de Vries et al., 2012; Amouri et al., 2014]. Most of the founder mutations were identified after extensive sequencing efforts and haplotype analysis of populations with a high prevalence of disease, and are primarily SNV and INDELs. The identification of founder pathogenic variants has facilitated the screening for carriers of these variants among unaffected individuals within these select populations. To identify and confirm whether a large deletion could be a founder mutation requires precise determination of the deletion breakpoints at the single-nucleotide level, which is not possible using the MLPA method.
Knowing the exact molecular changes that lead to disease may be critical for clinical management of the patient, especially for reproductive counseling. We have initiated application of high-throughput, state-of-the-art methodologies to identify all disease-causing variants in ∼300 FA families enrolled in the International Fanconi Anemia Registry (IFAR). As a part of this effort, we initially screened 202 FA families for deletions, using comparative genomic hybridization arrays (aCGH) and single-nucleotide polymorphism (SNP) arrays. We designed custom aCGH for simultaneous screening of deletions in 15 FA and other functionally relevant genes (the 16th FA gene, FANCQ, was recently identified and is thus not included in our array design). The arrays were designed to cover the entire length of each gene plus up to 200 kb on either side, enabling us to determine the precise boundaries of deletions extending beyond an FA gene locus. We identified deletions in 98 FA families in four different FA genes, of which 90% were in the FANCA gene. The high-resolution analysis of the deletion boundaries identified by aCGH, accompanied by subsequent cloning and sequencing of the breakpoints, provides insight into the location and potential mechanisms driving the intrachromosomal breakage events, in addition to identifying conserved deletions and their likely origin.
Materials and Methods
Study Subjects
Genomic DNA samples were from individuals diagnosed with FA and registered in the IFAR, following written informed consent. These studies were approved by the Institutional Review Board of the Rockefeller University, New York. The Office of Human Subjects Research at the National Institutes of Health and Institutional Review Board of the National Human Genome Research Institute (NHGRI) approved the reception of deidentified cell lines and DNA samples from The Rockefeller University and the analysis of the underlying molecular variants. Genomic DNA was isolated from peripheral blood, fibroblasts, or EBV-immortalized cell lines. The Puregene Kit and the DNeasy Blood and Tissue DNA Extraction Kit (Qiagen, Inc. Germantown, MD) were used for DNA extraction from blood and cell lines, respectively. Phenol/chloroform extraction and ethanol precipitation was included as a final step in the preparation of DNA.
aCGH
A custom CGH 12×135K array was designed using NimbleDesign (NimbleGen, Madison, WI). It consisted of 134,490 50 mer probes (44,830 probes in triplicates) selected with an interval of 37 bp. Unique probes were selected. The design covered the entire length and up to 200 kb on both sides of all the FA genes (except the recently identified FANCQ) and 12 other functionally relevant genes (Supp. Table S1). DNA from patients and reference DNA (human male DNA from Promega, Madison, WI) were labeled with different fluorochromes, mixed and hybridized to the 12 × 135,000 array. We used NimbleGen Service for CGH and thus the manufacturing, hybridization, scanning, and preliminary analysis were performed at their processing facility in Iceland. The data analysis was performed using NimbleScan and the intensity variations were visualized and displayed using SignalMap; both softwares were developed by NimbleGen. The genomic coordinates are based on the human genome build hg18 (NCBI36.1).
SNP Array
Genotyping was performed using the HumanCytoSNP, HumanOmniExpress, or HumanOmni2.5Quad DNA Analysis BeadChip Kit (Illumina, Inc., San Diego, CA) representing 300,000, 750,000 and 2.5 million SNPs, respectively. Genomic DNA (300 ng) was processed as per the Illumina “infinium assay” protocol [Gunderson et al., 2005]. In brief, this included whole-genome amplification and fragmentation of DNA, hybridization to the BeadChip with specific oligonucleotide probe array (50-mers), enzymatic extension of the 3′ terminal base for incorporation of the allele specific nucleotide, detection with fluorescently tagged reagent, and signal amplification. The allele type and its intensity were collected using iScan, and visualized with the GenomeStudio (v2011.1; www.Illumina.com) genotyping module. The deletion intervals in two families with larger deletions were determined using the intensity data using the same program.
PCR, Cloning, and Sequencing of Breakpoints
Multiple primer sets were designed (500–2,000 bp) flanking the breakpoint ends as determined by aCGH or SNP array. Amplification reactions were carried out with 10–15 ng of genomic DNA using KOD Extreme (EMD Millipore, Billerica, MA) enzyme according to the manufacturer recommendations with the addition of GC melt (Clonetech, Mountain View, CA). Initial 15 cycles of PCR, each with 0.5°C decreasing annealing temperature per cycle starting at 65°C was followed by 25 cycles at 57°C annealing temperature. Amplification products were analyzed by agarose gel electrophoresis, and the chosen products were purified from gel using Qiaquick gel extraction kit (Qiagen, Inc.). Purified products were A-tailed with Taq polymerase and cloned using TOPO TA-cloning kit (Life Technologies, Grand Island, NY). Clones were screened by colony PCR; plasmid DNA was purified from positive clones using Qiaprep Spin Miniprep Kit (Qiagen, Inc.), and subjected to cycle sequencing using BigDye v3.1 (Applied Biosystems, Grand Island, NY). Sequences were aligned to the human reference genome build hg18 (NCBI36.1) using the BLAT program (www.genome.ucsc.edu).
Haplotype Analysis
Genotypes of the proband and their parental DNA were collected using high-density SNP arrays as described above. Haplotypes associated with the FANCA-conserved deletions, CD2, and CD4–6, and the FANCC conserved deletion were determined using SNP genotypes from parental and proband DNA and analyzing the trios data with the -P1 option of PHASE v2.1 program (http://www.stat.washington.edu/Stephens/softwar.html) [Stephens et al., 2001; Stephens and Donnelly, 2003]. The output from this analysis along with knowledge of inheritance of each deletion allowed for the identification of the deleted allele. For FANCA, SNPs were chosen from chr16.hg18:g.88,235,597_88,513,344, and for FANCC, chr9.hg18:g.96,874,394_97,142,804. Only the SNPs that were also present in the HapMap Phased haplotype data (CEU, NCBI_Build36 rel22, http://www.Hapmap.org) were considered for analysis.
Phylogenetic Analysis
MEGA 5.1 (www.megasoftware.net) was used to perform a phylogenetic analysis on the FANCA common deletions. Phased haplotypes from the HapMap CEU population (Phase 2) were downloaded for use in the analysis as described above. We identified 51 unique CEU haplotypes for this genomic region as defined above. Of the 51 unique CEU haplotypes, four were excluded because each matched one of the seven haplotypes of the FANCA common deletions. The analysis consisted of 55 sequences: 47 unique haplotypes from the HapMap CEU population, seven haplotypes from the common deletions, and one from Chimp genome. The phylogeny reconstruction was performed using a maximum-likelihood statistical method with 500 bootstrap replications. A Tamura–Nei substitution model was implemented with the assumption that the rates were uniform for all sites (as opposed to gamma distributed). Missing data were not evaluated and are indicated with a question mark.
Results
aCGH Analysis Reveals Deletions in FANCA, FANCB, FANCC, and FANCD2
Genomic DNA from 202 FA families enrolled in the IFAR was screened for deletion variants by aCGH. The families chosen for analysis include 105 families with no prior screening for mutations, and 97 families for which one (69 families) or both mutations (28 families) were previously identified by various molecular methods including sequencing for SNV and INDELs and MLPA for larger deletions. Of the latter 97 families, 50 families (27 families with one mutation known and 23 families with both mutations known) were believed to carry deletions as determined by MLPA; however, precise breakpoint coordinates were lacking. Five families, for which both mutations were known from prior sequencing efforts, were used as negative controls for the array studies.
Genomic DNA samples from probands were screened for deletions for the entire length and up to 200 kb on either side of 15 FA genes, plus 12 additional functionally relevant genes (Supp. Table S1) using aCGH. Deletion variants were found in 88 FANCA, one FANCB, seven FANCC, and two FANCD2 families (Table 1A; Supp. Fig. S1). Two deletions, one FANCA and the FANCB, extended beyond the boundaries of the array design, and thus their deletion intervals were determined using genome-wide SNP arrays. For 48 of the 50 families previously thought to carry a deletion variant, as determined by other methods, deletions were confirmed and breakpoints defined; however, two single-exon deletions originally observed by MLPA were not confirmed by CGH, and subsequent sequence analysis revealed that sequence variants in these exons reduced hybridization of MLPA probes, thus giving an erroneous result in the initial MLPA assay. Of the 105 FA families for which there was no prior knowledge of the FA gene or the pathogenic variants, deletions accounted for one of the germline variants in 34 families (32 in FANCA and two in FANCD2) and both pathogenic variants in two families (both in FANCA, one homozygous family and one compound heterozygous family) (Table 1B). Thirty six of these 105 FA families carried a total of 38 deletions, indicating nearly a third of FA families carry deletions, and that the deletions constitute ∼18% of the total FA mutations (Table 1B).
| Gene | Deletion type | Families with deletion |
|---|---|---|
| A. All 202 FA families | ||
| FANCA | Heterozygous | 78 |
| Homozygous | 5 | |
| Compound heterozygous | 5 | |
| FANCB | Hemizygous | 1 |
| FANCC | Heterozygous | 7 |
| FANCD2 | Heterozygous | 2 |
| B. The subset of 105 FA families with no prior knowledge of affected gene or mutations | ||
| FANCA | Heterozygous | 32 |
| Homozygous | 1 | |
| Compound heterozygous | 1 | |
| FANCD2 | Heterozygous | 2 |
Characterization of FANCA Deletions in 88 FA Families
While 78 families with deletions in FANCA were heterozygous for the deletion, 10 families carried two deletions each; five probands carried distinct homozygous deletions, FAM12, FAM24, FAM26, FAM74, and FAM81 and the other five were compound heterozygotes resulting from two different deletions, FAM23, FAM28, FAM46, FAM47, and FAM63 (Table 1; Fig. 1). The two deletions in the compound heterozygote families overlapped with each other with the exception of FAM23, and parental DNA was screened to confirm the ends of the overlapping deletions when necessary. In total, 93 distinct FANCA-deleted alleles were identified, counting the deletions in homozygous families as one allele. The extent of the FANCA deletions is shown in Figure 1. The deletions’ sizes span a wide range from ∼1 to 545 kb, with over half of the deletions falling between 5 and 30 kb in length. The telomeric boundary of the largest deletion (FAM9) extended outside the design limits of the aCGH, and analysis using SNP array determined the deletion extended to the telomeric end of chr16. The deletions encompass as little as one exon within FANCA (FAM24, FAM30, FAM87, and FAM42) or as large as the entire FANCA gene along with 18 additional neighboring genes FAM1 (Fig. 1; Tables 2 and 3). Deletion data were deposited in the FA Mutation Database (http://www.rockefeller.edu/fanconi/).
| Family ID | Number of deletions | Gene | Chr | Deletion start | Deletion end | BPs | Deleted exons |
|---|---|---|---|---|---|---|---|
| 1 | 1 | FANCA | 16 | 88,131,700 | 88,610,500 | 478,801 | 1–43 |
| 3 | 1 | FANCA | 16 | 88,184,848 | 88,336,349 | 151,502 | 38–43 |
| 5 | 1 | FANCA | 16 | 88,227,499 | 88,446,249 | 218,751 | 1–43 |
| 6 | 1 | FANCA | 16 | 88,235,249 | 88,351,749 | 116,501 | 31–43 |
| 9a | 1 | FANCA | 16 | 88,281,756 | 88,827,254 | 545,499 | 1–43 |
| 12 | 2 HO | FANCA | 16 | 88,311,999 | 88,394,999 | 83,001 | 9–43 |
| 16 | 1 | FANCA | 16 | 88,330,849 | 88,500,000 | 169,152 | 1–43 |
| 20 | 1 | FANCA | 16 | 88,334,499 | 88,393,249 | 58,751 | 10–38 |
| 22 | 1 | FANCA | 16 | 88,338,249 | 88,470,249 | 132,001 | 1–36 |
| 41 | 1 | FANCA | 16 | 88,358,548 | 88,368,349 | 9,802 | 22–28 |
| 42 | 1 | FANCA | 16 | 88,358,574 | 88,359,274 | 701 | 28 |
| 43 | 1 | FANCA | 16 | 88,364,999 | 88,388,499 | 23,501 | 12–23 |
| 61 | 1 | FANCA | 16 | 88,383,499 | 88,474,299 | 90,801 | 1–14 |
| 62 | 1 | FANCA | 16 | 88,383,499 | 88,437,499 | 54,001 | 1–14 |
| 63_d1 | 2 HE | FANCA | 16 | 88,383,649 | 88,386,149 | 2,501 | 13–14 |
| 64 | 1 | FANCA | 16 | 88,386,749 | 88,401,249 | 14,501 | 7–11 |
| 65 | 1 | FANCA | 16 | 88,387,000 | 88,412,000 | 25,001 | 1–11 |
| 66 | 1 | FANCA | 16 | 88,387,749 | 88,432,749 | 45,001 | 1–11 |
| 69 | 1 | FANCA | 16 | 88,398,749 | 88,420,249 | 21,501 | 1–7 |
| 70 | 1 | FANCA | 16 | 88,398,749 | 88,425,749 | 27,001 | 1–7 |
| 72 | 1 | FANCA | 16 | 88,399,249 | 88,412,249 | 13,001 | 1–7 |
| 84 | 1 | FANCA | 16 | 88,403,105 | 88,410,000 | 6,896 | 2–5 |
| 85 | 1 | FANCA | 16 | 88,403,999 | 88,437,249 | 33,251 | 1–5 |
| 86 | 1 | FANCA | 16 | 88,404,000 | 88,405,000 | 1,001 | 4–5 |
| 88 | 1 | FANCA | 16 | 88,409,549 | 88,411,749 | 2,201 | 1–2 |
| 89 | 1 | FANCC | 9 | 96,935,249 | 96,937,749 | 2,501 | 8 |
| 96a | 1 | FANCB | X | 14,331,665 | 14,872,323 | 540,659 | 1–10 |
| 97 | 1 | FANCD2 | 3 | 10,048,249 | 10,066,749 | 18,501 | 2–17 |
| 98 | 1 | FANCD2 | 3 | 10,066,800 | 10,071,613 | 4,814 | 18 |
- This table excludes deletion intervals that were defined by cloning and sequencing, which are presented in Table 3.
- a Breakpoints for deletion determined by SNP array.
- Families with two deleted alleles are indicated with 2 in “Number of deletions” column.
- HO, homozygous.
- HE, compound heterozygous (and distinguished by “_d1” for deletion 1).
- The deletion coordinates are according to the NCBI36/hg18 build.
| FAM ID | No.∆ | FA gene | ∆Start | ∆ End | ∆Size (bp) | Cen rep | Cen rep start | Cen rep end | +/− | Tel rep | Tel rep start | Tel rep end | +/− | Overlap or insertion (bp)a | Potential mechanism | Alu--Alu | ∆Exons |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 1 | A | 88,172,538 | 88,337,600 | 165,063 | AluSx | 88,172,387 | 88,172,690 | + | AluSp | 88,337,451 | 88,337,757 | + | 11 | NAHR | Y | 37–43 |
| 4 | 1 | A | 88,224,066 | 88,408,991 | 184,926 | NR | AluSx | 88,408,947 | 88,409,245 | − | 3 | alt-EJ | N | 3–43 | |||
| 7 | 1 | A | 88,250,100 | 88,425,360 | 175,261 | AluY | 88,250,056 | 88,250,345 | − | AluY | 88,425,297 | 88,425,605 | − | 27 | NAHR | Y | 1–43 |
| 8 | 1 | A | 88,257,445 | 88,409,236 | 151,792 | AluSp | 88,257,167 | 88,257,453 | − | AluSx | 88,408,947 | 88,409,245 | − | 26 | NAHR | Y | 3–43 |
| 10 | 1 | A | 88,285,490 | 88,362,122 | 76,633 | NR | NR | 2 | alt-EJ | N | 27–43 | ||||||
| 11 | 1 | A | 88,302,909 | 88,483,387 | 180,479 | NR | NR | 4 | alt-EJ | N | 1–43 | ||||||
| 13 | 1 | A | 88,322,270 | 88,505,839 | 183,570 | AluSx | 88,322,116 | 88,322,301 | − | AluY | 88,505,703 | 88,506,000 | − | 24 | NAHR | Y | 1–43 |
| 14 | 1 | A | 88,327,695 | 88,377,962 | 50,268 | NR | AluSp | 88,377,678 | 88,377,968 | − | 2 | alt-EJ | N | 16–43 | |||
| 15 | 1 | A | 88,329,020 | 88,355,305 | 26,286 | AluSx | 88,328,786 | 88,329,049 | + | AluY | 88,355,047 | 88,355,357 | + | 11 | NAHR | Y | 30–43 |
| 17 | 1 | A | 88,331,201 | 88,352,999 | 21,799 | AluSg | 88,331,149 | 88,331,450 | + | AluY | 88,352,948 | 88,353,258 | + | 4 | NAHR | Y | 30–43 |
| 18 | 1 | A | 88,331,357 | 88,351,875 | 20,519 | AluSg | 88,331,149 | 88,331,450 | + | AluSx | 88,351,789 | 88,352,065 | − | 2 | alt-EJ | YO | 31–43 |
| 19 | 1 | A | 88,331,930 | 88,415,141 | 83,212 | NR | AluSx | 88,415,082 | 88,415,384 | − | 2 | alt-EJ | N | 1–43 | |||
| 21 | 1 | A | 88,336,484 | 88,414,112 | 77,629 | AluY | 88,336,437 | 88,336,568 | + | AluSx | 88,414,066 | 88,414,360 | + | 36 | NAHR | Y | 1–37 |
| 23_d1 | 2 HE | A | 88,341,384 | 88,375,356 | 33,973 | AluSg | 88,341,341 | 88,341,647 | + | AluY | 88,375,302 | 88,375,614 | + | 21 | NAHR | Y | 18–33 |
| 24 | 2 HO | A | 88,344,051 | 88,347,742 | 3,692 | AluJ/FLAM | 88,343,990 | 88,344,068 | + | AluSx | 88,347,682 | 88,347,981 | + | 31 | NAHR | Y | 31 |
| 25 | 1 | A | 88,344,194 | 88,355,131 | 10,938 | AluY | 88,344,109 | 88,344,414 | + | AluY | 88,355,047 | 88,355,357 | + | 7 | NAHR | Y | 30–31 |
| 26 | 2 HO | A | 88,344,637 | 88,404,186 | 59,550 | AluSq | 88,344,416 | 88,344,697 | + | AluSx | 88,403,786 | 88,403,949 | + | 17 | NAHR | Y | 6–31 |
| 27 | 1 | A | 88,345,159 | 88,389,286 | 44,128 | AluJb | 88,345,131 | 88,345,394 | + | AluSx | 88,389,257 | 88,389,566 | + | 10 | NAHR | Y | 12–31 |
| 28_d1 | 2 HE | A | 88,345,766 | 88,460,603 | 114,838 | AluY | 88,345,678 | 88,345,977 | − | AluY | 88,460,509 | 88,460,817 | - | 45 | NAHR | Y | 1–31 |
| 29 | 1 | A | 88,347,917 | 88,433,278 | 85,362 | AluSx | 88,347,682 | 88,347,981 | + | AluY | 88,432,969 | 88,433,298 | + | 11 | NAHR | Y | 1–30 |
| 30 | 1 | A | 88,348,428 | 88,353,975 | 5,548 | AluSq | 88,348,275 | 88,348,577 | − | NR | Insertion 37 bp | FoSTeS/MMBIR | N | 30 | |||
| 31 | 1 | A | 88,351,156 | 88,359,359 | 8,204 | AluSx | 88,350,890 | 88,351,168 | − | NR | Insertion 29 bp | FoSTeS/MMBIR | N | 28–30 | |||
| 32 | 1 | A | 88,355,680 | 88,365,679 | 10,000 | AluJb | 88,355,542 | 88,355,833 | − | FANCA exon23 | 88,365,587 | 88,365,723 | 2 | alt-EJ | N | 23–29 | |
| 33 | 1 | A | 88,356,841 | 88,368,468 | 11,628 | AluY | 88,356,875 | 88,357,188 | + | AluY | 88,368,358 | 88,368,663 | + | 21 | NAHR | Y | 22–28 |
| 34 | 1 | A | 88,356,984 | 88,370,562 | 13,579 | AluY | 88,356,875 | 88,357,188 | + | AluY | 88,370,455 | 88,370,769 | + | 44 | NAHR | Y | 21–28 |
| 35 | 1 | A | 88,356,998 | 88,373,127 | 16,130 | AluY | 88,356,875 | 88,357,188 | + | AluYINS | AluYINS | 88,373,127 | + | Insertion 179 bp AluY | FoSTeS/MMBIR | Y | 19–28 |
| 36 | 1 | A | 88,357,692 | 88,376,135 | 18,444 | AluSg | 88,357,566 | 88,357,859 | + | AluY | 88,376,003 | 88,376,138 | + | 4 | NAHR | Y | 18–28 |
| 37 | 1 | A | 88,357,692 | 88,376,135 | 18,444 | AluSg | 88,357,566 | 88,357,859 | + | AluY | 88,376,003 | 88,376,138 | + | 4 | NAHR | Y | 18–28 |
| 38 | 1 | A | 88,357,692 | 88,376,135 | 18,444 | AluSg | 88,357,566 | 88,357,859 | + | AluY | 88,376,003 | 88,376,138 | + | 4 | NAHR | Y | 18–28 |
| 39 | 1 | A | 88,357,733 | 88,365,259 | 7,527 | AluSg | 88,357,566 | 88,357,859 | + | AluJb | 88,365,086 | 88,365,390 | + | 20 | NAHR | Y | 18–24 |
| 40 | 1 | A | 88,357,923 | 88,376,192 | 18,270 | AluSq | 88,357,872 | 88,358,166 | + | AluY | 88,376,003 | 88,376,138 | + | 12 | NAHR | Y | 18–28 |
| 44 | 1 | A | 88,366,653 | 88,394,866 | 28,214 | AluSx | 88,366,445 | 88,366,750 | + | AluSx | 88,394,658 | 88,394,971 | + | 10 | NAHR | Y | 9–22 |
| 45 | 1 | A | 88,366,653 | 88,394,866 | 28,214 | AluSx | 88,366,445 | 88,366,750 | + | AluSx | 88,394,658 | 88,394,971 | + | 10 | NAHR | Y | 9–22 |
| 46_d1 | 2 HE | A | 88,368,466 | 88,404,057 | 35,592 | AluY | 88,368,358 | 88,368,663 | + | AluY | 88,403,951 | 88,404,249 | + | 28 | NAHR | Y | 6–21 |
| 47_d1 | 2 HE | A | 88,368,481 | 88,393,743 | 25,263 | AluY | 88,368,358 | 88,368,663 | + | AluY | 88,393,654 | 88,393,920 | + | 14 | NAHR | Y | 9–21 |
| 48 | 1 | A | 88,368,519 | 88,384,182 | 15,664 | AluY | 88,368,358 | 88,368,663 | + | AluSx | 88,384,029 | 88,384,350 | + | 8 | NAHR | Y | 15–21 |
| 49 | 1 | A | 88,368,578 | 88,375,523 | 6,946 | AluY | 88,368,358 | 88,368,663 | + | AluY | 88,375,302 | 88,375,614 | + | 14 | NAHR | Y | 18–21 |
| 50 | 1 | A | 88,368,840 | 88,373,628 | 4,789 | AluSx | 88,368,721 | 88,369,009 | + | NR | 2 | alt-EJ | N | 18–21 | |||
| 51 | 1 | A | 88,368,840 | 88,373,628 | 4,789 | AluSx | 88,368,721 | 88,369,009 | + | NR | 2 | alt-EJ | N | 18–21 | |||
| 52 | 1 | A | 88,372,488 | 88,396,714 | 24,227 | AluSg/x | 88,372,403 | 88,372,566 | + | LINE | 88,396,612 | 88,397,058 | + | None | NHEJ | N | 9–20 |
| 53 | 1 | A | 88,375,333 | 88,377,211 | 1,879 | AluY | 88,375,302 | 88,375,614 | + | AluY | 88,377,178 | 88,377,485 | + | 13 | NAHR | Y | 16–17 |
| 54 | 1 | A | 88,375,502 | 88,389,110 | 13,609 | AluY | 88,375,302 | 88,375,614 | + | AluSx | 88,388,912 | 88,389,192 | + | 22 | NAHR | Y | 12–17 |
| 55 | 1 | A | 88,376,050 | 88,377,224 | 1,175 | AluY | 88,375,627 | 88,375,930 | + | AluY | 88,377,178 | 88,377,485 | + | 22 | NAHR | Y | 16–17 |
| 56 | 1 | A | 88,376,126 | 88,377,300 | 1,175 | AluY | 88,376,139 | 88,376,434 | + | AluY | 88,377,178 | 88,377,485 | + | 14 | NAHR | Y | 16–17 |
| 57 | 1 | A | 88,376,281 | 88,382,229 | 5,949 | AluY | 88,376,139 | 88,376,434 | + | AluSc | 88,382,090 | 88,382,369 | + | 10 | NAHR | Y | 15–17 |
| 46_d2 | 2 HE | A | 88,383,475 | 88,391,064 | 7,590 | AluJo | 88,383,349 | 88,383,647 | − | AluY | 88,390,786 | 88,391,090 | + | Insertion 2 bp | NHEJ | YO | 11–14 |
| 63_d2 | 2 HE | A | 88,383,475 | 88,391,064 | 7,590 | AluJo | 88,383,349 | 88,383,647 | − | AluY | 88,390,786 | 88,391,090 | + | Insertion 2 bp | NHEJ | YO | 11–14 |
| 58 | 1 | A | 88,383,475 | 88,391,064 | 7,590 | AluJo | 88,383,349 | 88,383,647 | − | AluY | 88,390,786 | 88,391,090 | + | Insertion 2 bp | NHEJ | YO | 11–14 |
| 59 | 1 | A | 88,383,475 | 88,391,064 | 7,590 | AluJo | 88,383,349 | 88,383,647 | − | AluY | 88,390,786 | 88,391,090 | + | Insertion 2 bp | NHEJ | YO | 11–14 |
| 60 | 1 | A | 88,383,475 | 88,391,064 | 7,590 | AluJo | 88,383,349 | 88,383,647 | − | AluY | 88,390,786 | 88,391,090 | + | Insertion 2 bp | NHEJ | YO | 11–14 |
| 47_d2 | 2 HE | A | 88,384,547 | 88,394,863 | 10,317 | AluSg | 88,384,358 | 88,384,675 | + | AluSx | 88,394,658 | 88,394,971 | + | 23 | NAHR | Y | 9–14 |
| 28_d2 | 2 HE | A | 88,390,697 | 88,406,009 | 15,313 | AluSg | 88,390,650 | 88,390,785 | + | AluY | 88,405,963 | 88,406,268 | + | 8 | NAHR | Y | 4–10 |
| 67 | 1 | A | 88,393,778 | 88,401,667 | 7,890 | AluY | 88,393,654 | 88,393,920 | + | AluY | 88,401,513 | 88,401,817 | + | 12 | NAHR | Y | 7–8 |
| 68 | 1 | A | 88,394,840 | 88,404,129 | 9,290 | AluSx | 88,394,658 | 88,394,971 | + | AluY | 88,403,951 | 88,404,249 | + | 20 | NAHR | Y | 6–8 |
| 71 | 1 | A | 88,399,194 | 88,406,309 | 7,116 | FANCA exon 7 | 88,399,189 | 88,399,301 | − | AluSp | 88,406,274 | 88,406,579 | + | 1 | NHEJ | N | 4–7 |
| 73 | 1 | A | 88,399,898 | 88,409,021 | 9,124 | AluSq | 88,399,826 | 88,400,121 | − | AluSx | 88,408,947 | 88,409,245 | − | 5 | NAHR | Y | 3–6 |
| 74 | 2 HO | A | 88,400,101 | 88,419,559 | 19,459 | AluSq | 88,399,826 | 88,400,121 | − | AluY | 88,419,273 | 88,419,580 | − | 26 | NAHR | Y | 1–6 |
| 75 | 1 | A | 88,401,663 | 88,433,153 | 31,491 | AluY | 88,401,513 | 88,401,817 | + | AluY | 88,432,969 | 88,433,298 | + | 13 | NAHR | Y | 1–6 |
| 76 | 1 | A | 88,401,663 | 88,433,153 | 31,491 | AluY | 88,401,513 | 88,401,817 | + | AluY | 88,432,969 | 88,433,298 | + | 13 | NAHR | Y | 1–6 |
| 77 | 1 | A | 88,401,663 | 88,433,153 | 31,491 | AluY | 88,401,513 | 88,401,817 | + | AluY | 88,432,969 | 88,433,298 | + | 13 | NAHR | Y | 1–6 |
| 78 | 1 | A | 88,401,663 | 88,433,153 | 31,491 | AluY | 88,401,513 | 88,401,817 | + | AluY | 88,432,969 | 88,433,298 | + | 13 | NAHR | Y | 1–6 |
| 79 | 1 | A | 88,401,663 | 88,433,153 | 31,491 | AluY | 88,401,513 | 88,401,817 | + | AluY | 88,432,969 | 88,433,298 | + | 13 | NAHR | Y | 1–6 |
| 80 | 1 | A | 88,402,939 | 88,435,730 | 32,792 | AluY | 88,402,820 | 88,403,114 | − | AluSp | 88,435,611 | 88,435,932 | − | 13 | NAHR | Y | 1–5 |
| 81 | 2 HO | A | 88,403,094 | 88,418,774 | 15,681 | AluY | 88,402,820 | 88,403,114 | − | AluSg | 88,418,482 | 88,418,795 | − | 27 | NAHR | Y | 1–5 |
| 82 | 1 | A | 88,403,094 | 88,418,774 | 15,681 | AluY | 88,402,820 | 88,403,114 | − | AluSg | 88,418,482 | 88,418,795 | − | 27 | NAHR | Y | 1–5 |
| 83 | 1 | A | 88,403,104 | 88,418,467 | 15,364 | AluY | 88,402,820 | 88,403,114 | − | AluSc | 88,418,349 | 88,418,478 | − | 46 | NAHR | Y | 1–5 |
| 87 | 1 | A | 88,406,084 | 88,408,690 | 2,607 | AluY | 88,405,963 | 88,406,268 | + | AluSg | 88,408,570 | 88,408,879 | + | 22 | NAHR | Y | 3 |
| 23_d2 | 2 HE | A | 88,406,326 | 88,414,712 | 8,387 | AluSp | 88,406,274 | 88,406,579 | + | AluSq | 88,414,671 | 88,414,970 | + | 31 | NAHR | Y | 1–3 |
| 90 | 1 | C | 97,046,452 | 97,052,040 | 5,589 | NR | NR | Insertion 18 bp | FoSTeS/MMBIR | N | 2–3 | ||||||
| 91 | 1 | C | 97,046,452 | 97,052,040 | 5,589 | NR | NR | Insertion 18 bp | FoSTeS/MMBIR | N | 2–3 | ||||||
| 92 | 1 | C | 97,046,452 | 97,052,040 | 5,589 | NR | NR | Insertion 18 bp | FoSTeS/MMBIR | N | 2–3 | ||||||
| 93 | 1 | C | 97,046,452 | 97,052,040 | 5,589 | NR | NR | Insertion 18 bp | FoSTeS/MMBIR | N | 2–3 | ||||||
| 94 | 1 | C | 97,046,452 | 97,052,040 | 5,589 | NR | NR | Insertion 18 bp | FoSTeS/MMBIR | N | 2–3 | ||||||
| 95 | 1 | C | 97,115,943 | 97,125,170 | 9,228 | NR | MLT1J MaLR | 97,125,086 | 97,125,394 | + | Insertion 1 bp | NHEJ | N | 1 |
- a The precise sequence of the Overlap or Insertion bps is provided in Table S4
- Footnotes in Table 2 also apply here. Cen and Tel represent centromeric and telomeric ends of a deletion.
- The ∆Start is the Cen breakpoint and contains any overlap sequence.
- The ∆End coordinate is the Tel breakpoint and contains none of the overlap sequence.
- Strand (+/−) indicates the orientation of the repeat element.
- YO, yes in opposite orientation.
- Identical deletions are shaded in gray.
- The criteria for “potential mechanism” are as described in Yang et al. (2013).
- The deletion coordinates are according to the NCBI36/hg18 build.
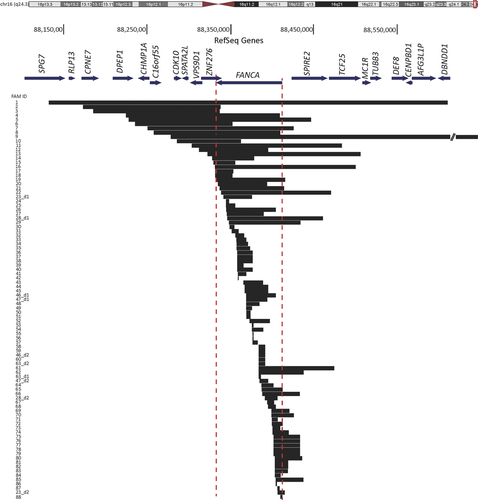
Fifty-two deletion ends were found to originate or terminate outside the boundaries of the FANCA gene, including 20 deletions starting centromeric to FANCA and 32 deletions ending telomeric to FANCA (Fig. 1). Eight of these deletions have both ends extending beyond the FANCA gene. In total, 47% of the deletions extend beyond the limits of the FANCA gene and the majority of these also affect other neighboring genes (Supp. Table S2). Similarly, analysis of the FANCA deletions from the subset of 105 FA families (Table 1B) for which no mutations were known prior to this study revealed 19 of the 35 FANCA deletions or 54% extend beyond FANCA.
Cloning, Sequencing, and Analysis of 68 FANCA Deletion Breakpoints
To further characterize the deletion breakpoints to the exact nucleotide, the region across the breakpoint junctions was amplified by designing multiple primer sets to a 500–2,000 bp region flanking the deletions. The resulting amplification products were purified, cloned, and sequenced. The primers used for cloning and sequencing of the breakpoints are listed in Supp. Table S3. Alignment of the sequences to the reference human genome identified the exact nucleotide positions of the breakpoints for 68 FANCA deletions. The results are summarized in Table 3 and the alignments are shown in Supp. Figure S2.
The extent of homology and the intervening sequences near the breakpoints reveal the likely mechanism(s) that cause the deletions, which are presented in Table 3 and Supp. Table S4 and summarized in Table 4. We find that majority (50/68) of the deletions have both breakpoints occurring at Alu elements in the same orientation with significant homology and overlapping sequence ranging from 4 to 45 bp. These appear to be driven by Alu–Alu-mediated nonallelic homologous recombination (NAHR) [Gu et al., 2008]. Two deletions occur at Alu elements in opposite orientation, one shared in FAM46_d2, FAM63_d2, and FAM58–60 and the other in FAM18. The former lacks any overlapping sequence but instead has a 2-bp insertion at the breakpoints, and thus appears to be mediated by nonhomologous end joining (NHEJ), whereas the latter has overlapping sequence of 2 bp and may be mediated by alternative end joining (alt-EJ). Nine of the 12 remaining breakpoints lack any significant homology surrounding the breakpoint junctions; FAMs 52 and 71 show 0–1 bp overlap and are anticipated to be driven by NHEJ, whereas the deletions in the other seven (FAMs 4, 10–11, 14, 18–19, and 32) with 2–4 bp homology appear to be caused by alt-EJ [Gu et al., 2008; Kidd et al., 2010; Verdin et al., 2013]. The alignment of breakpoint sequences to the reference genome of the final three deletions, FAMs 30, 31, and 35, indicate a complex rearrangement potentially involving more than two genomic regions. All three deletions contain inserted sequences of 37, 29, and 179 bp, respectively, between the fully aligned breakpoint junctions (Supp. Fig. S2; Supp. Table S4). The 37-bp insertion in FAM30 aligns 100% in the antisense orientation at chr16.hg18:g.88350637_88350672, which is located within the deleted interval. The 29-bp insertion in FAM31 could not be aligned to the genome reference sequence, and its origin is unclear. The 179-bp insertion in FAM35 is of AluY origin. This Alu element is not present in reference genome at the breakpoint coordinates but BLAT aligns with high homology (99%) approximately 2 kb downstream from the telomeric breakpoint at chr16.hg18:g.88,373,127. Due to unavailability of parental DNA, we were unable to confirm whether this is a de novo insertion of an Alu element or an inherited event. Breakpoint junctions with insertions of >10 bp, as observed in FAMs 30, 31, and 35, are thought to be caused by a replication-based mechanism termed FoSTeS/MMBIR (fork stalling and template switching/microhomology mediated break induced repair) [Lee et al., 2007].
| Gene | Proposed mechanisma | Nonrecurrent | Recurrent | Percentage | Observed breakpoint characteristics |
|---|---|---|---|---|---|
| FANCAb | FoSTeS/MMBIR | 3 | 0 | 5 | Insertions of 29, 34, and 178 bases |
| NHEJ | 2 | 1 | 5 | Homology of zero to one base or insertion of one to two bases | |
| alt-EJ | 7 | 1 | 15 | Homology of two, three, or four bases | |
| NAHR | 37 | 4 | 75 | Alu–Alu repeats in the same orientation | |
| FANCC | FoSTeS/MMBIR | 0 | 1 | – | Insertions of 18 bases |
| NHEJ | 1 | 0 | – | Insertion of one base |
- a Criteria for determining mechanism (as described in Yang et al., 2013).
- b The 68 FANCA deletions accounted for 55 distinct breakpoints
- NHEJ deletion breakpoints have an insertion (1–10 bp) or homology (<2 bp).
- FoSTeS/MMBIR deletion breakpoints have an insertion (>10 bp).
- alt-EJ deletion breakpoints do not have an insertion but have a homology (2–100 bp).
- NAHR deletion breakpoints do not have an insertion but have a homology (>100 bp).
Conserved Deletions in FANCA and Their Haplotype Analysis
aCGH indicated six deletions conserved in two or more seemingly unrelated families and sequence analysis confirmed these observations (Table 3; Fig. 2B; Supp. Table S5). CD2 (FAM44 and FAM45), CD1 (FAM36–38), and CD5 (FAM75–79) shared among two, three, and five families appeared to be caused by Alu–Alu-mediated NAHR. CD6 was similar in three families, FAM81–83: sequencing however confirmed it was identical in FAM81 and FAM82 but the deletion in FAM83 shared the same AluY element at the centromeric breakpoint with FAM81 and FAM82 but targets an alternate AluSc element at its telomeric breakpoint. NAHR is the likely origin of CD6 deletions as well. However, CD3 (FAM50 and FAM51) with 2 bp homology and CD4 (FAM46_d2, FAM63_d2, and FAM58–60) with 2 bp insertion appear to be caused by alt-EJ and NHEJ, respectively.
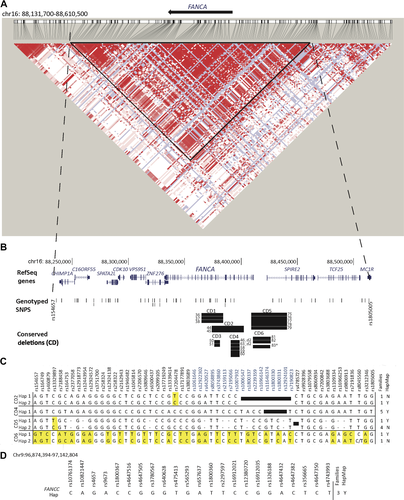
We performed SNP analysis on the proband and parental DNAs to identify the haplotype for the allele with the deletion (referred to as the deleted allele) for families with CD2, CD4, CD5, and CD6. Due to lack of parental DNA, we were unable to collect SNP data and thus generate haplotypes for the families with CD1 and CD3. Using the PHASE program, we were able to confidently establish the haplotype for the deleted alleles in families with CD2, CD4, CD5, and CD6. Results are summarized in Figure 2. For CD4, all five families were found to carry an identical haplotype on their respective deleted allele and support the hypothesis that this conserved mutation is probably the result of an ancient event. CD2 and CD5 each were identified on two very similar haplotypes, differing by just one SNP, and are also likely to be an ancient event. For CD2 and CD5, one of the haplotypes from each was found in the HapMap-phased haplotypes and the other was not. CD6 was found in two families on two very different haplotypes. This deletion therefore likely occurred independently in these two families (FAM81 and FAM82), thereby highlighting the breakpoints as potential hot spots. In support of this, the families are from different ethnic backgrounds, Caucasian and Hispanic (Supp. Table S5). In addition, FAM83 contains a very similar deletion interval as CD6, differing by just a few base pairs in the same Alu element at the centromeric end and a few hundred base pairs in an adjacent Alu element at the telomeric end of the deletion (Table 3). The similar deletions in FAM81 and FAM83 occur on the same haplotype and this haplotype is also found in the HapMap-phased haplotype database.
FANCA Deletion Breakpoints Reside Predominantly in AluY Small Interspersed Nuclear Elements
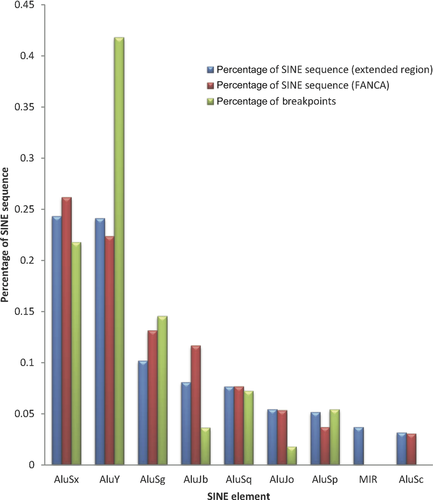
Distribution of the types of Alu at the FANCA locus and for 150 kb of the extended region encompassing the FANCA gene, and the extent of breakpoints in each Alu type is shown in Figure 3. AluSx and AluY are equally represented as the highest percentage of small interspersed nuclear element (SINE) sequence, and the equal distribution of AluSx and AluY is not limited solely to the FANCA gene but to the extended 150 kb region as well. However, AluY elements have 40% more breakpoints than AluSx elements (Fig. 3).
Locations of the deletion breakpoints and SINE elements within and around the FANCA gene are displayed in Figure 4. Despite the high number of available Alu elements in this region, some elements are targeted in multiple unique deletion events. Considering each conserved deletion as one deletion event that was caused by an ancient allele, with the exception of CD6, which may have occurred twice, there are 20 Alu elements involved in two or more deletions (Fig. 4; Supp. Table S6).

Deletions in FANCB, FANCC, and FANCD2 Families
Three distinct deletions were identified in FANCC (Supp. Fig. S1A; Tables 2 and 3). Five families, FAM90–FAM94, share the deletion eliminating exons 2–3. Sequence analysis confirmed the shared deletion and an 18-bp insertion at the breakpoint junction, suggesting the deletion is likely caused by a fork stalling and template switching/microhomology mediated break-induced replication (FoSTeS/MMBIR) mechanism (Table 3; Supp. Fig. S2). Haplotype analysis could be performed for three of the five families carrying the conserved deletion (Fig. 2D), and the three families shared the same haplotype, which supports the idea that deleted alleles are likely the result of a common ancestral mutation. The other two deletions, one encompassing exon one and regions upstream of the gene and one encompassing exon 8, were each found in one family, FAM95 and FAM89, respectively. The deletion breakpoints in FAM95 mapped precisely to the reference genome and had a “C” residue inserted at the breakpoint junction, suggesting NHEJ causing the deletion.
The heterozygous deletions in two FANCD2 families were intragenic eliminating exon 18 (FAM98) or exons 2–17 (FAM97), whereas the hemizygous deletion in the FANCB family (FAM96) removed the entire gene and was large, requiring use of SNP array to determine the deletion boundaries (Table 2; Supp. Fig. S1 B and C).
Discussion
Knowing the molecular changes that lead to a disease is necessary for the advancement of “personalized” genomics and precision medicine [Couch et al., 2014]. The techniques we describe here can be employed toward precise molecular diagnosis of FA, which offers an increased potential for establishing genotype–phenotype relationships and translating these relationships to aid in management of the disease. The current study of deletions is part of our comprehensive effort to identify all the pathogenic variants for FA families in the IFAR. Recognizing that deletions contribute substantially to the mutation spectrum in FANCA, and that FANCA is mutated in 60% of FA patients, FANCA deletion analysis by MLPA was proposed as an initial step in a comprehensive mutation screening strategy earlier [Ameziane et al., 2008]. However, MLPA has primarily been limited to the identification of deleted FANCA exons and thus there has been very little effort to expand the screening for all FA genes at once and to define the precise molecular nature of the deletions.
Nearly a Third of FA Patients Carry Deletions, and Half of the FANCA Deletions Extend Beyond the Gene
Our study included screening for deletions in a subset of 105 families for which there was no prior data on the affected gene or the mutations causing the disease, and thus provided an evaluation of the extent of deletions in FA. We observed a third of these families carrying deletions in FA genes. Though predominantly in FANCA, deletions were also observed in other FA genes, indicating that screening for deletions in all FA genes as described here is an invaluable tool in the molecular diagnosis of FA patients. The aCGH, though designed for high-resolution scan, may not reveal very small deletions; however, the lower limit is not clear. Our array design that allowed for scanning for deletions up to 200 kb beyond the boundaries of FA genes, and precise mapping of the deletions allowed us to determine that half of the FANCA deletions extended beyond the boundaries of the gene. Thus, the MPLA method, which is limited to detection of deleted FANCA exons, is insufficient to fully characterize almost half the deletions in FANCA. The two largest deletions eliminated the largest number of surrounding genes in addition to the entire FANCA gene: the 545 kb in FAM9 eliminated 16 additional genes, four centromeric and 12 telomeric; the 478-kb deletion in FAM1 eliminated 18 additional genes, 10 centromeric and eight telomeric (Fig. 1; Supp. Table S2). The large hemizygous deletion (541 kb) in the FANCB family removed the entire FANCB as well as the neighboring GLRA2 gene (Supp. Fig. S1C). Identification of a larger number of such extended deletions would enable a reliable evaluation of whether the haploinsufficiency (or complete loss) caused by the elimination of a subset of neighboring genes affects phenotypic heterogeneity or influences the clinical outcome of FA patients. As observed for a FANCC deletion [Chandrasekharappa et al., 2013], the deletions removing the regulatory regions of FANCA may effect expression of the deletion-carrying allele, which can now be evaluated using a series of precise deletions identified in this study.
Three Quarters of the FANCA Deletions Appear to Have Originated by NAHR
The identification of the exact nucleotide breakpoints provides insight into the mechanisms driving the deletion events in FA genes. Potential mechanisms leading to structural variants have been revealed, such as in the recent analysis of >2,000 breakpoint junctions in the human genome [Kidd et al., 2010] and somatic structural variations in human cancer genome from 140 patients [Yang et al., 2013]. We observed that 75% of the FANCA deletions are Alu–Alu-mediated NAHR, whereas NHEJ (5%), alt-EJ (15%), and FoSTeS/MMBIR (5%) account for the rest (Table 4). Alu–Alu-mediated NAHR is one of the most prevalent mechanisms driving recurrent intrachromosomal recombination events in genomic disorders caused by deletions [Gu et al., 2008; Liu et al., 2011], and it seems to be the predominant mode for generating FANCA nonrecurrent deletions as well. Alu–Alu-mediated NAHR has been suggested to be the cause for majority of deletions in Von Hippel–Lindau (VHL) [Franke et al., 2009] and in BRCA1 [Mazoyer, 2005]. A majority of the nonrecurrent deletions in the FOXL2 gene region leading to the blepharophimosis syndrome, however, lacked extended homology and were found to involve mechanisms other than NAHR [Verdin et al., 2013]. About 20% of the FANCA deletions share no homology or very short homology (1–4 bp), and the mechanism driving these deletions may be explained by NHEJ (or alt-EJ), an imprecise DNA repair mechanism for double-stranded breaks that does not require homology and tolerates addition of nucleotides at the joining sites [Weterings and van Gent, 2004; Lieber, 2008]. Interestingly, one FANCC and three FANCA deletions with insertions of >10 bp at the breakpoints appear to be caused by FoSTeS/MMBIR, a replication-based mechanism that involves stalling at replication forks and switching to a different template, and thus may involve more than two regions. In fact, two of the three FANCA deletions appear to involve three regions, reminiscent of a recent description of an Alu-mediated deletion involving SOX10 regulatory elements associated with Wardenburg syndrome type 4 [Bondurand et al., 2012]. The precise mapping of the breakpoints as described here for gene regions with multiple deletions will enhance our understanding of the origin of pathogenic variants.
Hotspots for Deletion Breakpoints in AluY SINE Elements at FANCA Introns 5, 17, and 21
The identification of the precise nucleotide breakpoint by sequencing across the breakpoint junctions confirmed that the majority of breaks in FANCA occur in Alu elements (122/136). This is not surprising given the high density of SINE within the FANCA locus. Genome-wide Alu SINE elements account for 11% of the total human genome sequence; however, at the FANCA locus, SINE elements comprise nearly 40% of the total genomic sequence. The genome-wide distribution of various Alu subfamilies reveals AluS followed by the oldest Alu element, AluJ, are the most abundant, accounting for 6.4% and 2.5% of the total genome sequence, respectively. The youngest of the Alu subfamilies, AluY [Price et al., 2004], is the least abundant subfamily at 1.5%; however, the distribution throughout the genome appears to be nonrandom [Grover et al., 2003]. AluSx and AluY are equally represented as the highest percentage of SINE sequence at the FANCA locus and for 150 kb of the extended region encompassing the FANCA gene (Fig. 3). Thus, the equal distribution of AluSx and AluY is not limited solely to the FANCA gene in this region. Despite the fact that the AluSx and AluY elements are of equal abundance across the FANCA region, the breakpoints preferentially occur in AluY elements. Indeed, AluY elements have 40% more breakpoints than AluSx elements, highlighting that AluY elements are preferred for Alu-mediated NAHR in FANCA (Fig. 3). The increased homology among the AluY elements may favor them as predominant sites for intrachromosomal deletions. A study analyzing the deletion breakpoints associated with VHL disease, also reported that an AluY element was involved in seven out of 33 deletions of the VHL gene region, and thus highly recombinogenic [Franke et al., 2009].
Despite the high number of available Alu elements in this region, some elements are targeted in multiple unique deletion events. Alu elements with multiple breakpoints may represent potential hotspots for DNA breakage events in FANCA, particularly those containing breakpoints from four or more unique deletions (Fig. 4; Supp. Table S6). We also identified clusters of Alu elements with multiple breakpoints at introns 5, 17, and 28 of FANCA; the highest density of breakpoints occurred at intron 17 with nine unique breakpoints within a 1 kb region. It is not clear why certain Alu elements are targeted multiple times or why there is a high density of breakpoints in certain locations within the FANCA gene. It does not appear to be solely dependent on the density of Alu elements, nor is there any discernible correlation with DNase hypersensitivity sites, which might indicate accessible DNA. It is possible that a higher order of chromatin structure and overall genomic architecture may influence the availability and proximity of elements for recombination events.
Our analysis of conserved deletions identified six deletions in FANCA and one deletion in FANCC that are identical in two or more seemingly unrelated families of full or partial Caucasian/European descent. The FANCC mutation discovered earlier by RNA analysis, and denoted as c.1–250del because it lacked the first 250 bp from cDNA [Strathdee et al., 1992], is in fact caused by this genomic deletion, encompassing exons 2–3. However, one conserved deletion (CD6) was found on two very different haplotypes from families of different ancestry, supporting its occurrence at two different times. Phylogenetic analysis of the deleted haplotypes with the HapMap CEU Phase 2 haplotypes reveals that CD2, CD4, CD5, and CD6.2 are relatively closer to each other. With the exception of CD6.1, the deleted alleles appear to be of very recent origin (Supp. Fig. S3).
Although the majority of the FANCA deletions were unique and private, the high-resolution CGH data revealed that many of the deletions might share a common breakpoint at one or both ends. Six deletions appear to have near-identical breakpoint junctions shared in two or more families. Conservation of the breakpoint events may indicate hotspots for chromosomal breakage within or around the FANCA gene. Our haplotype analysis of the families with conserved deletions indicates that except for CD6 that arose independently in two families, the rest appear to be acquired through shared inheritance.
aCGH is an integral component of a comprehensive strategy for identifying disease-causing variants in FA genes [Chandrasekharappa et al., 2013]. Our analysis here reveals the extent and the broad spectrum of deletions in multiple FA genes that contribute to the onset of FA. Efforts can now be made to discover the distinct pathogenic molecular events caused by these variants, and to correlate these with associated phenotypic changes. The identification of precise breakpoints allows for quick screening of deletions in family members by PCR-based methods and provides insight into the mechanisms driving deletion mutations.
Acknowledgments
We acknowledge the support from the Intramural Program of the National Human Genome Research Institute, NIH, Bethesda, MD. E.K.F. acknowledges an NIH FARE award. We thank Marypat Jones and Ursula Harper of the Genomics Core, NHGRI for performing SNP genotyping. A.S. is a Rita Allen Foundation, Irma T. Hirschl, and Alexandrine and Alexander Sinsheimer Foundation scholar, and is a recipient of a Doris Duke Clinical Scientist Development Award. Finally, we are most grateful to the individuals and families who participated in this study, and for the help of their physicians in enrolling them in the IFAR.
Disclosure statement: The authors declare no conflict of interest.




