Major Histocompatibility Complex Class I-Related Chain A Alleles and Histology of Nonalcoholic Fatty Liver Disease
Supported by the Beatty Liver and Obesity Research Program, Inova Fairfax Hospital and Inova Health System Seed (Grant IRB 12.401 to Z.M.Y.).
Potential conflict of interest: Nothing to report.
Abstract
Major histocompatibility complex class I-related chain A (MICA) is a highly polymorphic gene that modulates immune surveillance by binding to its receptor on natural killer cells, and its genetic polymorphisms have been associated with chronic immune-mediated diseases. The progressive form of nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), is characterized by accumulation of fat and inflammatory cells in the hepatic parenchyma, potentially leading to liver cell injury and fibrosis. To date, there are no data describing the potential role of MICA in the pathogenesis of NAFLD. Therefore, our aim was to assess the association between MICA polymorphism and NASH and its histologic features. A total of 134 subjects were included. DNA from patients with biopsy-proven NAFLD were genotyped using polymerase chain reaction–sequence-specific oligonucleotide for MICA alleles. Liver biopsies were assessed for histologic diagnosis of NASH and specific pathologic features, including stage of fibrosis and grade of inflammation. Multivariate analysis was performed to draw associations between MICA alleles and the different variables; P ≤ 0.05 was considered significant. Univariate analysis showed that MICA*011 (odds ratio [OR], 7.14; 95% confidence interval [CI], 1.24-41.0; P = 0.04) was associated with a higher risk for histologic NASH. Multivariate analysis showed that MICA*002 was independently associated with a lower risk for focal hepatocyte necrosis (OR, 0.24; 95% CI, 0.08-0.74; P = 0.013) and advanced fibrosis (OR, 0.11; 95% CI, 0.02-0.70; P = 0.019). MICA*017 was independently associated with a higher risk for lymphocyte-mediated inflammation (OR, 5.12; 95% CI, 1.12-23.5; P = 0.035). Conclusion: MICA alleles may be associated with NASH and its histologic features of inflammation and fibrosis. Additional research is required to investigate the potential role of MICA in increased risk or protection against NAFLD.
Abbreviations
-
- ALT
-
- alanine aminotransferase
-
- AST
-
- aspartate aminotransferase
-
- BMI
-
- body mass index
-
- CD
-
- cluster of differentiation
-
- CI
-
- confidence interval
-
- HCC
-
- hepatocellular carcinoma
-
- HDL
-
- high-density lipoprotein
-
- HLA
-
- human leukocyte antigen
-
- HSC
-
- hepatic stellate cell
-
- MAF
-
- minor allele frequency
-
- MFI
-
- mean fluorescence intensity
-
- MICA
-
- major histocompatibility complex class I-related chain A
-
- NAFLD
-
- nonalcoholic fatty liver disease
-
- NASH
-
- nonalcoholic steatohepatitis
-
- NK
-
- natural killer
-
- NKG2D
-
- natural killer cell group 2, member D
-
- OR
-
- odds ratio
-
- PBMC
-
- peripheral blood mononuclear cell
-
- PCR
-
- polymerase chain reaction
-
- RTU
-
- ready to use
Nonalcoholic fatty liver disease (NAFLD) is a chronic progressive form of liver disease that encompasses a wide spectrum. The clinical and histopathologic spectrum ranges from steatosis without nonalcoholic steatohepatitis (NASH), which is NAFLD, to NASH, advanced fibrosis, and cirrhosis.(1) The exact mechanisms that are implicated in the pathogenesis of NAFLD and NASH remain ill-defined and poorly understood. Current evidence suggests a “multiple-hit hypothesis” that incorporates an interplay of lipid accumulation, lipotoxicity, oxidative stress, necrotic–inflammatory processes, and genetic factors.(2) The role of immunogenetic markers potentially involved in the development and progression of this disease has been suggested in recent years. Particular emphasis has been placed on the role of the human leukocyte antigen (HLA) allele, although another important locus may be the major histocompatibility complex (MHC) class I-related chain A (MICA). Initial investigations carried out on HLA alleles showed that HLA-B65, HLA-DQB1, and HLA-DQ5 were expressed at a considerably higher level in patients with NAFLD and were correlated with NASH and severe liver fibrosis outcomes.(3, 4) Another study suggested that both HLA class I and II gene polymorphisms are involved in the pathogenesis of this disease.(5)
Unlike the classical MHC class I and II molecules and HLA alleles, studies have shown that patients with chronic liver disease have elevated levels of MICA in their sera. As part of the innate arm of the immune system, MICA is a membrane glycoprotein ligand expressed more commonly on transformed rather than normal cells.(6) The presence of this ligand indicates its role in coordinating a stress-mediated signal transduction response through the natural killer (NK) cell group 2, member D (NKG2D) receptor present on NK cells, cluster of differentiation (CD)8+ T cells, and γδ T cells.(6, 7) A preliminary study investigating MICA in patients with NASH undergoing bariatric surgery showed enhanced levels, supporting its potential role in this liver disease.(7) Further, MICA genetic mechanisms play a potential role in autoimmunity. In other liver diseases, such as autoimmune hepatitis (AIH), MICA may play an important role. In a study conducted by Oliveira et al.,(8) the MICA*08 allele was more common in patients with AIH with the DRB1*03 allele. Additionally, MICA may be important in the pathogenesis of hepatic malignancy and nonhepatic fibrotic diseases. Zhang et al.(9) showed that hepatic tissues from patients with hepatocellular carcinoma (HCC) had reduced expression and associated this reduction with advanced stages of HCC, suggesting its potential role in tumor aggressiveness.
In non-liver fibrotic disease, MICA has also been implicated in idiopathic pulmonary fibrosis (IPF). In fact, immunoreactivity of MICA was significant, as indicated by alveolar epithelial cells and fibroblasts being positive in patients with IPF compared to controls.(10) Finally, anti-MICA antibodies may play a role in both graft rejection and poor graft survival rates in solid organ transplants.(11, 12)
With its role as an important immunogenetic regulator in the host and among various chronic multisystem diseases, the MICA gene necessitates further exploration. The overall aim of our study was to investigate the association between MICA gene polymorphisms and NAFLD and its histologic features. We conducted this study by comparing MICA alleles in a NAFLD cohort of patients with obesity and undergoing bariatric surgery and a cohort of control patients who were not known to have NAFLD.
Patients and Methods
Study Design
This study cohort comprised 134 patients enrolled from 2010 to 2012. The patients in the study included biopsy-proven NAFLD (n = 81) and healthy controls without liver disease (n = 53) (Table 1). Blood samples, demographic features, and clinical data were obtained at the time of liver biopsy after a comprehensive informed consent process. Liver biopsies were obtained during bariatric surgery for a clinical indication for suspected liver disease and were read by our histopathologist. Only patients 18 years of age or older were enrolled, and exclusion criteria included other liver disease, a history of alcohol intake (>10 g/day for women and 20 g/day for men), and history of pregnancy. The control group was enrolled from blood bank donors who were known to be healthy and with no known history of chronic disease, although we were not able to assess them specifically for the presence of NAFLD because there was no indication for liver biopsy. Control participants were not available for assessment of liver steatosis by ultrasound or any noninvasive blood tests.
| Variable | NAFLD (n = 81) | Control (n = 53) | P Value |
|---|---|---|---|
| Demographic features | |||
| Age (years), mean (SD) | 43.74 (10.66) | 55.09 (16.05) | <0.001 |
| BMI (kg/m2), mean (SD) | 44.88 (9.22) | 27.06 (4.58) | <0.001 |
| Male, n (%) | 17 (21.0%) | 31 (59.6%) | <0.0001 |
| White, n (%) | 58 (71.6%) | 50 (94.3%) | <0.01 |
| Variable | Non-NASH NAFLD (n = 49) | NASH NAFLD (n = 32) | P Value |
|---|---|---|---|
| Demographic features | |||
| Age (years), mean (SD) | 42.49 (10.65) | 45.66 (10.56) | 0.21 |
| BMI (kg/m2), mean (SD) | 45.21 (9.10) | 44.36 (9.53) | 0.73 |
| Male, n (%) | 5 (10.2%) | 12 (37.5%) | 0.003 |
| White, n (%) | 32 (65.3%) | 26 (81.3%) | 0.12 |
| Clinical features | |||
| Diabetes, n (%) | 16 (32.7%) | 12 (37.5%) | 0.65 |
| Hyperlipidemia, n (%) | 28 (59.6%) | 19 (61.3%) | 0.88 |
| Hypertension, n (%) | 24 (52.2%) | 19 (59.4%) | 0.53 |
| Albumin (g/dL), mean (SD) | 4.09 (0.54) | 4.10 (0.62) | 0.77 |
| Total bilirubin (mg/dL), mean (SD) | 0.45 (0.20) | 0.82 (1.21) | 0.002 |
| Alkaline phosphatase (U/L), mean (SD) | 88.80 (35.09) | 87.74 (24.18) | 0.71 |
| AST (U/L), mean (SD) | 23.93 (11.67) | 36.06 (26.40) | 0.005 |
| ALT (U/L), mean (SD) | 32.50 (18.93) | 51.71 (34.33) | 0.003 |
| Glucose (mg/dL), mean (SD) | 117.80 (55.39) | 127.93 (56.21) | 0.45 |
| Total cholesterol (mg/dL), mean (SD) | 200.86 (57.53) | 195.17 (38.53) | 0.97 |
| LDL (mg/dL), mean (SD) | 110.15 (33.49) | 115.35 (29.03) | 0.55 |
| HDL (mg/dL), mean (SD) | 47.88 (15.22) | 40.00 (7.49) | 0.009 |
| Triglycerides (mg/dL), mean (SD) | 184.90 (115.58) | 192.03 (85.55) | 0.32 |
- P value shows results of the χ2 test for categorical variables and Mann-Whitney U test for continuous variables. P ≤ 0.05 is considered significant. Categorical data are shown as frequency (n) and percentage (%), and continuous variables are shown as mean ± SD.
- Abbreviation: LDL, low-density lipoprotein.
Samples in this study had sera separated, with peripheral blood mononuclear cells (PBMCs) snap frozen at −80°C for later DNA extraction. This study was not conducted until complete institutional review board approval was received from our human protection program at Inova Fairfax Hospital.
Liver Biopsy Histopathologic Evaluation for NAFLD
Liver biopsies were formalin fixed in preparation for sectioning and subsequent microscopic assessment. Each section was stained with hematoxylin and eosin to evaluate for NAFLD and Masson’s trichrome to evaluate for fibrosis. All biopsies were evaluated and read by one hepatopathologist (Z.G.). Each liver biopsy was assessed for histopathologic features, including the degree of steatosis (0-3; 0 is <5% fat, 1 is 5%-33% fat, 2 is 34%-66% fat, and 3 is >67% fat), portal inflammation (0-3), lymphoplasmacytic lobular inflammation (0-3), polymorphonuclear lobular inflammation (0-3), Kupffer cell hypertrophy (0-3), presence of apoptotic bodies (0-3), focal parenchymal necrosis (0-3), hepatocellular (degenerative) ballooning (0-3), Mallory-Denk bodies (0-3), interlobular pericellular fibrosis (0-3), portal fibrosis (0-3), bridging fibrosis (absent = 0; few = 1), and cirrhosis (absent = 0; incomplete = 1). Higher scores indicated an advanced progression of the histologic features. Correct NASH diagnosis was based on the minimal criteria of presence of hepatic steatosis, lobular inflammation, and ballooning of hepatocytes with or without pericellular fibrosis. The NAFLD activity score is designed to quantify activity and stage of fibrosis and was used accordingly.(13, 14)
MICA Genotyping
DNA Isolation From PBMCs
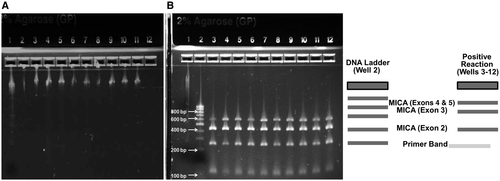
Isolation of DNA from 200 µL of PBMCs from both controls and patients was conducted using the QIAamp DNA Isolation Kit (QIAGEN, Germantown, MD), with strict adherence to the manufacturer’s protocol. Spectrophotometric analysis allowed for determination of the quality of the extracted DNA, with an absorbance (A)260/A280 ratio of 1.6 to 2.0 as a reference range for good quality. Additionally, agarose gel electrophoresis (Fig. 1A) enabled confirmation of DNA integrity. The eluted DNA was subsequently stored at −20°C until further analysis.
Polymerase Chain Reaction Sequence-Specific Oligonucleotide Method
MICA typing of patients followed the protocol described by the LABType sequence-specific oligonucleotide (SSO) kits from One Lambda-Thermo Fisher Scientific on the Luminex 100/200. Briefly, polymerase chain reaction (PCR) was used to amplify MICA alleles. For every PCR reaction, 2 µL of genomic DNA (20 ng/µL) was added to each of 96 wells in a PCR tray. The amplification mix consisted of 432 µL of ready-to-use (RTU) primer mix, 1,491 µL of D-mix, and 22 µL of Taq polymerase; 18 µL of the amplification mix (13.8 µL D-mix, 4 µL amplification primer, 0.2 µL Taq polymerase) was added to each well containing DNA. After being vortexed and centrifuged for 3 to 5 seconds, 18 µL of the master mix mixture was aliquoted into each well containing DNA. PCR samples were then amplified with the C1000 Bio-Rad thermal cycler in the following steps: 96°C for 3 minutes for 1 cycle; 96°C for 20 seconds, 60°C for 20 seconds, 72°C for 20 seconds for 5 cycles; 96°C for 10 seconds, 60°C for 15 seconds, 72°C for 20 seconds for 30 cycles; 72°C for 10: minutes for 1 cycle; 4°C forever for 1 cycle. Then, 5 µL of the amplified PCR sample was loaded onto a 2% agarose gel to verify the amplified product (Fig. 1B).
In a new tray, 5 µL of the amplified DNA sample was placed into a clean well and mixed with 2.5 µL of denaturation buffer until a bright pink color emerged. This mixture was incubated at room temperature for 10 minutes. Then, 5 µL of neutralization buffer was added to the mixture, and 38 µL of hybridization mixture (hybridization buffer + bead mixture), consisting of Luminex microspheres coated with specific probes for complementary sequences of 88 MICA alleles (MICA*001-MICA*088) was added to each well containing neutralized DNA. Additionally, in every reaction, there were three positive controls (bead 013, exon 2; bead 032, exon 3; bead 039, exons 4 and 5) and one negative control (bead 035). The PCR tray containing the beads was incubated in a 60°C thermal cycler for 15 minutes. The tray was washed and centrifuged at 1,000g with 100 µL of wash buffer 3 times.
We then added 50 µL of 1X R-phycoerythrin-conjugated-streptavidin (SAPE) (57.5 µL stock SAPE Reagent and 5,693 µL SAPE buffer) to each well to label PCR products. The tray was vortexed at low speed and incubated for 5 minutes in a 60°C thermal cycler and then washed once again. After removing the supernatant, a wash buffer was added to make the final volume 80 µL. All samples were mixed and transferred to a 96-well microplate for data, using the Luminex 100/200 instrument.
Results were analyzed by calculating the mean fluorescence intensity (MFI) generated by the Luminex Data Collector of each bead per sample as follows: percent positive value = 100 × MFI (probe n) − MFI (probe negative control) ÷ MFI (probe positive control) − MFI (probe negative control). Percent positive values were compared to predetermined cut-off values provided by the LABType SSO testing kit. Allele groups in each sample were determined by matching patterns of positive and negative beads based on information provided by the testing kit. The data were then analyzed by HLA Fusion 3.0 (One Lambda).
MICA and CD161 Immunohistochemical Staining of Liver Sections
A total of 26 formalin-fixed liver biopsies from patients with NAFLD in our cohort were deparaffinized using HistoChoice (#H103-4L; IHC World, Ellicott City, MD). Heat-induced antigen retrieval was performed using IHC-Tek Epitope Retrieval Solution (#IW-1100-1L; IHC World), followed by blocking with BLOXALL Endogenous Peroxidase and Alkaline Phosphatase Blocking Solution (#SP-6000; Vector Laboratories, Inc., Burlingame, CA) for 10 minutes and Streptavidin/Biotin Blocking Kit (#SP-2002; Vector Laboratories) for 10 minutes. Slides were washed using IHC-Tek RTU Washing Buffer Phosphate-Buffered Saline-Tween 20 (#IW-1200; IHC World). We then added 200 µL of anti-MICA antibody (3 µg/mL) (#ab62540; Abcam, Cambridge, United Kingdom) and anti-CD161 antibody (5 µg/mL) (#ab197979; Abcam), which were diluted in IHC-Tek Antibody Diluent (#IW-1000; IHC World), to each slide and incubated the slides at 4°C overnight, followed by washing. Then, RTU Biotinylated Universal Antibody (#BP-1400; Vector Laboratories) was added to each slide and incubated at room temperature for 45 minutes. After incubation, the slides were washed, and 1 drop of RTU VECTASTAIN Elite ABC Horseradish Peroxidase (HRP) reagent (#PK-7100; Vector Laboratories) was added to each slide for 30 minutes, followed by washing. Next we added 100 µL of ImmPACT DAB Peroxidase (HRP) substrate (#SK-4105; Vector Laboratories) to each slide and incubated the slides for 8 minutes, followed by washing. Finally, slides were counterstained with Vector Hematoxylin QS (#H-3404; Vector Laboratories) for 1 minute and washed. Slides were mounted using VectaMount Permanent Mounting Medium (#H-5000; Vector Laboratories).
Statistical Analysis
Clinical, demographic, and biochemical data of the study cohort groups were summarized as frequency, percentage, and mean ± SD. The Mann-Whitney U test was used for continuous variables, and the nonparametric χ2 test was used to compare categorical data and MICA alleles between disease states (NAFLD vs. controls, NASH NAFLD vs. non-NASH NAFLD, or fibrosis stage ≥2 vs. fibrosis stages 0-1).
Multivariate logistic regression models were presented as odds ratio (OR), with 95% confidence interval (CI) of MICA alleles used as predictors of disease outcomes (NAFLD vs. control, NAFLD vs. NASH) after adjustment for demographic, clinical, and biochemical confounders (age, sex, race, bilirubin, aspartate aminotransferase [AST], alanine aminotransferase [ALT], and high-density lipoprotein [HDL]). To limit the chance of overfitting, the analytes were preselected at the univariate stage (P < 0.10 by Mann-Whitney or Spearman’s correlation test), and only predictors with P ≤ 0.05 were left in the models after bidirectional stepwise selection. All statistical analyses were performed using statistical software JMP 9.2 (SAS Institute, Cary, NC).
Results
Demographic and Clinical Characteristics of Patients With and Without NASH
The study cohort comprised 134 patients in two groups: (1) biopsy-proven NAFLD (NASH [n = 32], age 45.7 ± 10.6 years, men 37.5%; non-NASH [n = 49], age 42.5 ± 10.6 years, men 10.2%), and (2) control ([n = 53], age 55.1 ± 16 years, men 59.6%). Patients with NASH (n = 32) had significantly higher levels of ALT (mean ± SD, 51.7 ± 34.3 U/L vs. 32.5 ± 18.9 U/L; P = 0.003) and AST (36.0 ± 26.4 U/L vs. 23.9 ± 11.7 U/L; P = 0.005) than patients without NASH (n = 49). Other demographic and clinical features are summarized in Table 1. A comparison of histopathologic features of NASH versus non-NASH are summarized in Table 2.
| Variable | Non-NASH NAFLD (n = 49) | NASH NAFLD (n = 32) | P Value |
|---|---|---|---|
| Histologic features | |||
| Advanced steatosis, n (%) | 6 (12.2%) | 16 (50.0%) | <0.001 |
| Lobular and portal inflammation | |||
| Inflammatory Kupffer cells, n (%) | 0 (0.0%) | 3 (11.1%) | 0.02 |
| Inflammatory lymphocytes, n (%) | 3 (6.1%) | 13 (40.6%) | <0.001 |
| Inflammatory PMN leukocytes, n (%) | 3 (6.1%) | 6 (18.8%) | 0.08 |
| Portal inflammation, n (%) | 23 (46.9%) | 22 (68.8%) | 0.05 |
| Apoptosis and necrosis | |||
| Mallory-Denk bodies, n (%) | 0 (0.0%) | 11 (34.4%) | <0.0001 |
| Focal necrosis, n (%) | 24 (49.0%) | 23 (71.9%) | 0.04 |
| Fibrosis | |||
| Pericellular fibrosis, n (%) | 0 (0.0%) | 30 (93.8%) | <0.0001 |
| Portal fibrosis, n (%) | 25 (51.0%) | 26 (81.3%) | 0.006 |
| NAS | |||
| NAS (>4) (%) | 3 (6.0%) | 21 (65.6%) | <0.0001 |
- The histopathologic scoring system by hepatopathologist Dr. Zachary Goodman classified these features into two groups: mild or moderate/severe. Steatosis (mild = 0-2, moderate/severe = 3-4); lymphoplasmacytic lobular inflammation (mild = 0-1, moderate/severe = 2-3); PMN lobular inflammation (mild = 0-1, moderate/severe = 2-3); portal inflammation (mild = 0-1, moderate/severe = 2-3); focal necrosis, apoptosis, and Mallory-Denk bodies (mild = 0, moderate/severe = 1-2); pericellular and portal fibrosis (mild = 0-1, moderate/severe = 2-3). NASH diagnosis was based on the minimal criteria of presence of hepatic steatosis, lobular inflammation, and ballooning of hepatocytes with or without pericellular fibrosis. NAS is a scoring designed to quantify activity and stage of fibrosis and was used accordingly.
- Abbreviations: NAS, nonalcoholic fatty liver disease activity score; PMN, polymorphonuclear.
MICA Allele Associations With NAFLD
In our cohort of 81 patients with NAFLD, the minor allele was MICA*002, observed 23 times with a minor allele frequency (MAF) of 0.142 (MAF/minor allele count MICA*002 = 0.142/23). In the cohort of 53 controls, the minor allele was also MICA*002, observed 15 times with a MAF of 0.142 (MAF/minor allele count MICA*002 = 0.142/15). Additionally, in both cohorts, MICA*008 was the most common allele, observed 42 times among patients with NAFLD (allele frequency = 0.259) and 31 times among controls (allele frequency = 0.292). An overall comparison for MICA allele frequencies among patients with NAFLD and control patients is shown in Supporting Table S1.
Univariate analysis revealed that MICA*007 was significantly less frequent in patients with NAFLD (4.9%) versus controls (15.1%) (P = 0.04). Similarly, MICA*012 was less frequent in NAFLD (1.2%) versus controls (9.4%) (P = 0.03). On the other hand, MICA*027 was significantly more frequent in patients with NAFLD (11.1%) versus controls (1.9%) (P = 0.047) (Fig. 2).
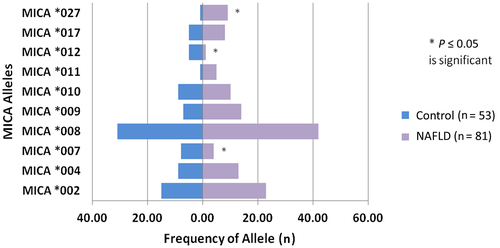
In the multivariate model after controlling for race, age, and sex, MICA*007 remained independently associated with a lower risk of NAFLD (OR, 0.12; 95% CI, 0.01-0.97; P = 0.047), whereas MICA*027 was independently associated with a higher risk for NAFLD (OR, 8.67; 95% CI, 1.03-73.2; P = 0.047) (Table 3). Analyses of these alleles are depicted in Fig. 2.
| Variable | OR (95% CI) | P Value |
|---|---|---|
| Age | 0.93 (0.90-0.96) | <0.001 |
| BMI | 1.50 (1.28-1.75) | <0.001 |
| Male | 0.08 (0.03-0.23) | <0.001 |
| White | 0.11 (0.03-0.42) | 0.001 |
| MICA*007 | 0.12 (0.01-0.97) | 0.047 |
| MICA*027 | 8.67 (1.03-73.2) | 0.047 |
- OR with 95% CIs obtained in the corresponding logistic regression model in which other significant variables were presented in the model.
MICA Allele Associations With NASH
In our cohort of 49 patients without NASH but with NAFLD, the minor allele was MICA*002, observed 17 times with a MAF of 0.173 (MAF/minor allele count MICA*002 = 0.173/17). In the cohort of 32 patients with NASH with NAFLD, the minor alleles were MICA*002, MICA*004, and MICA*009, each observed 6 times with a MAF of 0.094 (MAF/minor allele count MICA*002/MICA*004/MICA*009 = 0.094/6). Additionally, in both cohorts, MICA*008 was the most common allele, observed 25 times among patients without NASH but with NAFLD (frequency = 0.255) and 17 times among controls (frequency = 0.266). An overall comparison for MICA allele frequencies among patients with and without NASH is shown in Supporting Table S1.
MICA and CD161 Expression in Liver Tissue
Out of 23 patients with NAFLD, immunohistochemical staining of MICA revealed that most patients constitutively expressed MICA throughout the liver tissue, although to varying degrees, as exemplified by a liver-tissue staining from one of our patients with NASH (Supporting Fig. S1A). In 18 out of 23 patients, more than 30% of the liver-tissue area was positive for MICA staining. Immunohistochemical staining of anti-CD161 antibody showed localized expression of NK cells. Out of the 23 patients, most showed low CD161 expression (Supporting Fig. S1B).
Multivariate Analysis of MICA Alleles With NASH
Univariate analysis in this study showed that MICA*011 is associated with a higher risk for histologic NASH (OR, 7.14; 95% CI, 1.24-41; P = 0.03) (Fig. 3C).
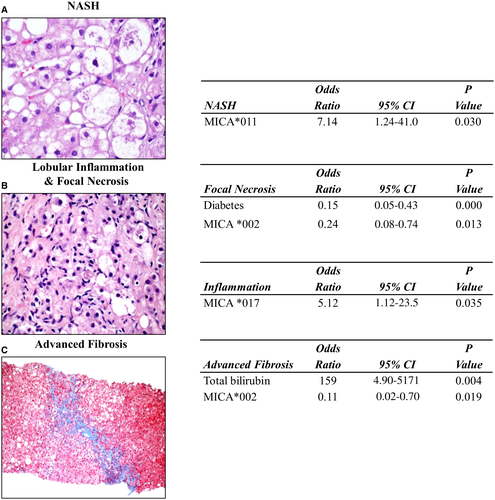
In the multivariate model, after controlling for sex and important biochemical markers (bilirubin, AST, ALT, and HDL), MICA*011 remained independently associated with a higher risk for NASH (P ≤ 0.05). However, because of the high CI, the significance of this result should not be overemphasized. No other MICA alleles were found to be significantly associated with a lower or higher risk of NASH in the study.
MICA Allele Associations With Specific Histologic Features of NASH
Univariate analysis in this study showed that MICA*002 is associated with a lower risk for focal necrosis (OR, 0.34; 95% CI, 0.12-0.92; P = 0.03) and advanced fibrosis (OR, 0.20; 95% CI, 0.04-0.92, P = 0.04). Furthermore, MICA*017 is associated with a higher risk for inflammation (OR, 5.08; 95% CI, 1.11-23.2; P = 0.04).
In the multivariate model, after controlling for sex and important biochemical markers (bilirubin, AST, ALT, and HDL), it was shown that MICA*002 remained independently associated with a lower risk for focal necrosis (OR, 0.24; 95% CI, 0.08-0.74; P = 0.013) as well as advanced fibrosis (OR, 0.11; 95% CI, 0.02-0.70; P = 0.019). Further results indicated that MICA*017 showed independent association with a higher risk for inflammation (OR, 5.12; 95% CI, 1.12-23.5; P = 0.035) (Fig. 3C).
Discussion
This study primarily focused on the potential role of MICA polymorphism in NAFLD and its subtypes (NASH and non-NASH) and known healthy controls. It is important to place some of our findings in the context of accumulating evidence suggesting the role of MICA in liver diseases, such as NAFLD and NASH. Although our current understanding of the pathogenic mechanisms of NAFLD is not precise, it is postulated that immunogenetic factors may be of critical significance.(15-18) Hepatic fibrogenesis involves a complex interplay among a wide variety of cells, with each one being activated by mediators responsible for subsequent tissue injury.(19) The culprit cells directly implicated in fibrosis are the hepatic stellate cells (HSCs), which on activation, proliferate and produce excessive extracellular matrix leading to the formation of the fibrotic scar.(19) Krizhanovsky et al.(20) have shown that higher levels of immune modulators, such as MICA, are present in senescent activated HSCs, which can act to limit fibrosis progression compared to proliferating activated HSCs. Moreover, the senescent cells are selectively targeted by NK cells, potentially through interaction of the MICA ligand and NKG2D receptor. Thus, identifying these MICA polymorphisms associated with NAFLD/NASH and histologic features of NAFLD/NASH may help to assess genetic predisposition to disease and elucidate potential molecular targets for therapy.
Our data showed that MICA*007 was associated with a lower risk for NAFLD in contrast to MICA*027, which was associated with a higher risk for NAFLD. MICA*011 remained independently associated with a higher risk for NASH. Interestingly, the study by Gong et al.(21) on the association of MICA gene polymorphisms with infectious liver fibrosis in patients with schistosomiasis showed that MICA*027 was more frequent in healthy controls, which renders the role of this allele controversial. In our study, patients with NAFLD with advanced liver fibrosis were found to have a significantly lower frequency of MICA*002 that also correlated negatively with focal necrosis. This may be explained by the fact that reduced engagement with the NKG2D receptor might lead to less activation of NK cells and subsequently to less cytotoxicity against activated HSCs and therefore more aggressive development of liver fibrosis. This may be supported further by the very low expression of CD161 (NK-cell marker) in liver tissues from our patients with NAFLD, indicating that NK cells are less activated. Additionally, MICA*017 remained independently associated with a higher risk for inflammation. MICA is recognized by γδ T cells, CD8+ T cells, and NK cells, which may trigger the cytologic and inflammatory responses of these cells. In fact, in a study by Allez et al.,(22) MICA–NKG2D interaction has been implicated in the inflammatory and cytotoxic responses in patients with Crohn’s disease. This suggests that CD4+ T cells expressing NKG2D could be important effector cells that influence the immune-mediated inflammatory process in NAFLD.
Several studies have also demonstrated that various MICA family members are elevated in the sera of patients with HCC.(23-25) Furthermore, we found that there was constitutive expression of MICA in the liver tissue of patients with and without NASH. MICA may play a role as a risk factor in those patients who will develop HCC. In fact, Huang et al.(26) showed that patients with hepatitis C virus with cirrhosis who carried MICA risk alleles or had high levels of serum MICA were at a higher risk for development of HCC after failed antiviral therapy. Further studies are needed to identify patients who are at risk for developing HCC in the NAFLD population. Additionally, knowledge of MICA risk alleles could be used to develop targeted therapies, as demonstrated by Wang et al.,(27) who developed an rG7S–MICA fusion protein that demonstrated successful antitumor activity in mouse models with HCC by recruiting NK cells to induce cytokine release.
Despite the importance of innate immunity in inflammatory liver diseases and the role of NK-cell activation in their pathogenesis, this study is one of the earliest addressing the role of MICA alleles in the pathogenesis of NAFLD. Although our patient cohort was limited, the central reading by a single hepatopathologist (Z.G.) and his in-depth analysis of histologic features in patients with biopsy-proven NAFLD are strengths of our study. Additionally, we used medium- to high-resolution MICA genotyping. However, a potential limitation of our cohort may be the significantly different body mass index (BMI), age, sex, and race between our control and NAFLD groups. Although age, sex, and race were controlled for in the multivariate model, we did not control for BMI in order to avoid our associations being influenced by the strong correlation between obesity and NAFLD. In our NAFLD cohort, all patients were morbidly obese (BMI >44 kg/m2); therefore, as expected, both subsets of patients with and without NASH have underlying conditions of obesity and metabolic syndrome (diabetes, hypertension, hyperlipidemia). It should also be noted that our study of MICA polymorphisms was conducted in a NAFLD cohort that consisted of all patients with obesity and bariatric surgery, so conclusions may not be generalizable to all patients with NAFLD. Additionally, although our healthy control group was selected from blood bank donors who had no known history of chronic diseases, we cannot rule out the presence of NAFLD in this population because of the high prevalence of NAFLD overall. In addition, control participants were unavailable for us to assess the presence of NAFLD using ultrasound or any noninvasive blood tests.
We initially used BMI as a predictor in our multivariate model; however, we realized that it had the same effect as NAFLD, noting that all patients with NAFLD were morbidly obese (BMI >44 kg/m2) compared to controls (BMI 27 kg/m2). Therefore, we did not include BMI as a predictor. However, to limit the chance of overfitting, the analytes were preselected at the univariate stage (P < 0.10 by Mann-Whitney or Spearman correlation test), and only predictors with P ≤ 0.05 were left in the models after bidirectional stepwise selection. This was done to reduce the chances of finding false-positive associations, especially in light of large CIs. Due to the limited sample size, we were only able to test for a limited number of hypotheses without substantially increasing the risk of overfitting. Although some associations were found between some of the MICA alleles and NAFLD/NASH, it is still premature to derive any direct causality. Future studies are needed to validate these potential associations in patients with NASH.
In summary, several MICA alleles were found to be associated with either increased risk or protection related to NAFLD/NASH, supporting the importance of the cytolytic T-cell response to the outcome of NASH and fibrosis. In this context, the immune repertoire of an individual might be implicated in NAFLD pathogenesis, and therefore cellular immunity needs to be further studied in patients with NAFLD and NASH. Of particular note, we found that 1) MICA*007 was significantly more frequent in patients without NAFLD and independently associated with a lower risk of NAFLD. Additionally, MICA*012 was less frequent in patients with NAFLD versus controls. 2) MICA*027 was significantly more frequent in patients with NAFLD compared to controls and independently associated with a higher risk for NAFLD. 3) MICA*011 was positively associated with higher risk for NASH. 4) Histologically, MICA*002 remained independently associated with a lower risk for focal necrosis and advanced liver fibrosis. 5) MICA*017 was independently associated with a higher risk for liver inflammation. 6) NAFLD is a heterogeneous disease comprising various biological and clinical subtypes. The immunogenetic markers identified in this study are valuable in providing a more coherent understanding of the mechanisms underlying histopathologic changes.
Acknowledgment
We thank Linda Henry for proofreading the manuscript.