Fibroblast Growth Factor 15–Dependent and Bile Acid–Independent Promotion of Liver Regeneration in Mice
Supported by the Rutgers Busch Grant and Rutgers Start-up Fund (R01GM104037 and BX002741 to G.L.G.; R01GM118376 to X.B.Z.).
Abstract
The role of intestine-derived factors in promoting liver regeneration after partial hepatectomy (PHx) are not entirely known, but bile acids (BAs) and fibroblast growth factor 15 (Fgf15) that is highly expressed in the mouse ileum could promote hepatocyte proliferation. Fgf15 strongly suppresses the synthesis of BAs, and emerging evidence indicates that Fgf15 is important for liver regeneration. The mechanisms by which Fgf15 promotes liver regeneration are unclear, but Fgf15 may do so indirectly by reducing BA levels and/or directly by promoting cell proliferation. However, it remains undetermined whether these two mechanisms are independent or integrated. In this study, we aimed to clarify these relationships by generating Fgf15 Tet-Off, transgenic mice (Fgf15 Tg) that had very low BA levels as a result from overexpressed Fgf15-mediated suppression of BA synthesis. Compared with wild-type mice, the Fgf15 Tg mice showed increased hepatocyte proliferation even without surgery, and a further induction of the genes in cell-cycle progression after PHx. Moreover, overexpression of Fgf15 by adeno-associated virus (AAV)-Fgf15 transduction or treatment with the recombinant Fgf15 protein led to increased cell proliferation in vivo. Furthermore, Fgf15 Tg mice exhibited an earlier and greater activation of mitogen-activated protein kinase, signal transducer and activator of transcription 3, and NF-κB signaling pathways in the priming stage, and a disruption of the hippo signaling pathway in the termination stage of liver regeneration. Conclusion: Direct in vivo evidence demonstrates that Fgf15 is critical in stimulating the phases of priming and termination of liver regeneration that are critical for cell survival and liver-size determination, independent of BA levels. (Hepatology 2018; 00:000-000).
The liver is capable of a full regeneration following partial resection or chemical-induced injury through a highly orchestrated process. Two-thirds partial hepatectomy (2/3 PHx) in rodents has been used widely as a model to study liver regeneration since its introduction,1 with the complex regenerative processes and a cohort of factors being well-characterized.2
Bile acids (BAs) have emerged as a group of critical factors in liver regeneration. BAs are synthesized in the liver from cholesterol, undergo enterohepatic circulation, and are indispensable to emulsify lipids and lipid-soluble vitamins for intestinal absorption. Moreover, BAs function as important physiological signaling molecules to activate nuclear receptors, including farnesoid X receptor (FXR), pregnane X receptor, and vitamin D receptor. Through their effects on these nuclear receptors, BAs regulate hepatic lipid, glucose, and energy homeostasis, and maintain metabolic balances.3, 4 BAs can also activate G protein-coupled receptors (GPCRs), including sphingosine 1-phosphate receptor (S1PR) 2 and G protein-coupled bile acid receptor 1 (also called TGR5), to regulate lipid and glucose homeostasis.5-7 Disorders in BA homeostasis have been implicated in cholestatic liver diseases, fatty liver disease, cardiovascular disease, obesity, and diabetes.4, 8-10
Fibroblast growth factor 15 (Fgf15) (human ortholog FGF19) is expressed in the ileum, and highly induced upon activation of FXR.11, 12 It is well known that Fgf15 functions as a gut-derived hormone to suppress BA synthesis through the Fgf15/Fgfr4 axis.13 Briefly, secreted Fgf15 interacts with Fgfr4/β-Klotho receptor complexes at the cell membrane of hepatocytes, which leads to activation of c-Jun N-terminal kinase (JNK) and extracellular signal-regulated kinase (ERK) pathways and ultimately the suppression of the expression of genes in BA synthesis, including Cyp7a1/CYP7A1 and Cyp8b1/CYP8B1.11, 13, 14
BAs have been shown to promote liver regeneration. Specifically, depletion of BAs by BA-sequestering resin (cholestyramine) resulted in a decrease in liver regeneration,15, 16 which is consistent with a report showing that external biliary drainage led to decreased liver regenerative capacity.17 Mechanistically, BAs promote liver regeneration by activating FXR either in the liver to directly induce cell cycle progression or in the intestine to induce Fgf15/FGF19 that activates hepatocyte proliferation and suppresses toxic BA levels.16, 18, 19 Indeed, the role of BA receptors in liver regeneration has been shown, and mice deficient in FXR or TGR5 displayed significantly higher mortality, severe liver necrosis, and delayed liver regeneration after PHx.16, 20-22
The intestine-derived Fgf15/FGF19 is important to liver growth. We and others have shown that Fgf15 and FGF19 are critical in promoting liver regeneration and defining hepatic size, and mice deficient in Fgf15 showed impaired liver regeneration, marked liver injury, and higher mortality.19, 21, 22 The underlying mechanisms by which Fgf15/19 promotes liver regeneration could be by reducing the BA levels to prevent BA-induced liver injury or through direct promotion of cell proliferation.21, 22 Therefore, we need to determine the direct effects of Fgf15 on liver regeneration versus the indirect mechanism regulated through suppressing BA synthesis. To determine the direct, BA-independent effects of Fgf15 on liver regeneration in vivo, we generated a transgenic (Tg) mouse model, with Fgf15 overexpression in both the liver and intestine using the tetracycline inducible Tet-Off system. This model allows us to determine the role of Fgf15 in liver regeneration in vivo—independent of BA levels.
Materials and Methods
ANIMALS AND TREATMENTS
Tissue-specific, inducible Fgf15 Tg mice were generated on the C57BL/6J background (Supporting Information). Whole-body Fgf15 knockout (KO) mice were described previously.22, 23 Male mice (n = 5-12/group) were used in this study, with age-matched C57BL/6J mice as wild-type (WT) controls. Mice were maintained in a pathogen-free animal facility at 22°C under the 12-hour light/dark cycle. The animal studies were approved by the Rutgers University Institutional Animal Care and Use Committee and comply with all relevant federal guidelines and institutional policies. Fgf15 Tg and WT mice were administered by oral gavage with water (10 mL/kg body weight [BW]) containing 2 mg/mL doxycycline (Dox, Sigma, D9891) daily for 5 days to stop the Fgf15 transgene expression. Two days after the treatment with Dox, 2/3 PHx was performed as described previously,22 with details in the Supporting Information. Adeno-associated virus (AAV)–mediated overexpression of Fgf15 was achieved in Fgf15 KO and WT mice through a single tail-vein injection of 1 × 1013 plaque-forming units/kg of AAV or AAV-Fgf15. Mice were kept in quarantine for 7 days before undergoing 2/3 PHx. Tissues were collected 48 hours after surgery. For cholic acid (CA) feeding, WT and Fgf15 Tg mice were fed chow diet containing 0% or 0.1% CA for 7 days. Recombinant Fgf15 protein (rFgf15) produced in E. coli and purified as described previously24, 25 was injected into mice through tail vein at a dosage of 50 μg/kg. Tissues were collected 3 hours after injection.
SERUM BIOCHEMICAL PARAMETER ANALYSIS
Serum activities/levels of alanine transaminase (ALT), aspartate transaminase (AST), Alkaline phosphatase (ALP), total triglycerides (TGs), total cholesterol (TC), and total bilirubin were determined by kits from Pointe Scientific (Canton, MI) and the total BA assay kit was from Diazyme Laboratories (Poway, CA).
DETERMINATION OF BA POOL SIZE
The total BA pool size was determined and expressed as micromoles of BAs per 100 g BW (μmol/100 g BW). The BA concentration in the liver was expressed as μmol/g liver weight (LW), as described previously13 and with details in the Supporting Information.
HISTOLOGICAL AND IMMUNOHISTOCHEMICAL ANALYSIS
Hematoxylin and eosin (H&E) staining and immunohistochemistry staining of Fgf15 and bromodeoxyuridine (BrdU) are described in detail in the Supporting Information.
ISOLATION OF RNA, REAL-TIME QUANTITATIVE PCR, AND WESTERN BLOT ANALYSIS
Samples were prepared and analyzed according to previous publications.13, 22 Gene expression at mRNA levels was normalized to the housekeeping gene, β-Actin, with primers listed in the Supporting Table S1. β-actin or α-tubulin protein levels were used as the loading control for western blot analysis.
STATISTICAL ANALYSIS
All experimental data are expressed as the mean ± SD. Statistical differences among multiple groups were tested using two-way analysis of variance followed by the Student-Newman-Keuls test. A value of P < 0.05 was considered statistically significant.
Results
GENERATION AND CHARACTERIZATION OF Fgf15 TG MICE
To determine the direct effect of Fgf15 on liver regeneration in vivo, a line of Fgf15 Tg mice were generated by using the Tet-Off expression system to create a tissue-specific expression of Fgf15 that can be turned off by Dox treatment (Fig. 1A). Specifically, two lines of mice were cross-bred to make the Fgf15 Tg mice, with one expressing tetracycline transactivator fusion protein (tTA) driven by the promoter of fatty acid binding protein (FABP) 1 gene for tissue-specific overexpression of tTA protein in both the liver and intestine,26, 27 and with the other line carrying a tTA-responsive element (TRE) consisting of a repeated tetracycline operator linked to the target gene of interest, herein Fgf15 (described in detail in the Supporting Information). Therefore, Fgf15 Tg mice have Fgf15 overexpression in both enterocytes and hepatocytes where the FABP1 promoter functions. Oral administration of Dox by gavage can efficiently prevent the binding of tTA protein to the TRE sequence, which prevents Fgf15 transgene overexpression. The time course of changes in Fgf15 mRNA levels following Dox treatment is shown in Supporting Fig. S1. Fgf15 Tg mice developed and reproduced normally. When the Tg mice reached 2 to 3 months of age, the serum activities/levels of ALT, AST, ALP, TGs, TC, BAs, and total bilirubin were measured. Despite elevated serum BA levels in Tg mice compared with WT mice, these levels were still within the normal range. All other serum parameters were similar between Tg and WT mice (Fig. 1B). Fgf15 Tg mice had lower BW and LW/BW ratio compared with WT mice (Fig. 1C). No significant difference in liver histology/pathology was observed between WT and Tg mice (Fig. 1D). In WT mice, Fgf15 protein was expressed in differentiated epithelial cells of the intestine and not detected in the livers, but in Fgf15 Tg mice, strong Fgf15 protein expression was detected in both periportal hepatocytes and intestinal epithelial cells (Fig. 1E).
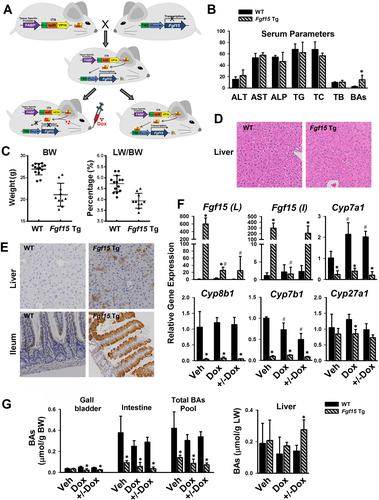
Levels of Fgf15 mRNA in Tg mice were 300-fold to 600-fold higher than those in WT mice (Fig. 1F). Treatment with Dox for 5 days terminated the Fgf15 overexpression in both liver and intestine, and interestingly, 5 days after Dox withdrawal, the expression of Fgf15 transgene was only recovered in the intestine but not the liver (+/−Dox, Fig. 1F), suggesting that this strategy could be used as an intestine-specific Fgf15 overexpression model. With overexpression either in both the liver and intestine (vehicle [Veh], Fig. 1F) or only in intestine (+/−Dox, Fig. 1F), Fgf15 Tg mice showed markedly reduced mRNA levels of Cyp7a1, Cyp8b1 and Cyp7b1, but not Cyp27a1 (Fig. 1F). The total BA pool size in Tg mice was 20% to 30% that in WT mice (Fig. 1G). Specifically, Fgf15 overexpression did not change BA levels in the liver, but reduced BA concentration in the intestines (Fig. 1G), where most of the BAs are stored in the body. To specifically determine the role of Fgf15 overexpression in the intestine, the Tg mice treated with Dox for 5 days were used for all of the subsequent experiments.
OVEREXPRESSION OF Fgf15 REGULATES GENES INVOLVED IN BA HOMEOSTASIS AND CELL PROLIFERATION WITH OR WITHOUT BA SUPPLEMENT
To restore their BA pool size, Fgf15 Tg mice were fed a 0.1% CA-containing diet (Fig. 2). With CA feeding, Tg mice showed a similar increase in liver BA levels compared with WT mice, but markedly increased BA levels in the intestine, and as a result, the total BA pool size increased in Tg mice (Fig. 2A). Hepatic Fgf15 mRNA levels were induced with CA feeding, with a significantly higher induction in Tg than in WT mice. Suppression of the gene expression of Cyp7a1, Cyp8b1, Cyp7b1, and Cyp27a1 was achieved with CA feeding, and further suppression of Cyp7a1 and Cyp8b1 was noted in Fgf15 Tg mice (Fig. 2B). Without CA feeding, Fgf15 Tg mice had lower mRNA levels of Shp compared with WT mice. The CA feeding reduced mRNA levels of Ntcp, but increased mRNA levels of Shp, Bsep, and Ostβ in WT mice (with the latter two not significant). However, CA feeding did not alter Ntcp and Shp mRNA levels in Fgf15 Tg mice, and markedly increased those of Bsep and Ostβ (Fig. 2B). In the ileum, the higher Fgf15 expression in Tg mice was associated with lower expression of Ibabp, which was highly induced by CA in both WT and Tg mice. Even though on chow diet the Ostβ expression was reduced in Fgf15 Tg mice compared with WT, CA feeding significantly increased Ostβ expression only in Fgf15 Tg mice (Fig. 2C).
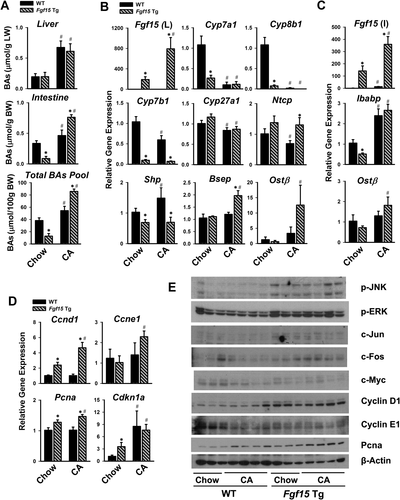
A previous study showed that BAs stimulate hepatocyte proliferation.16 Therefore, to determine to what extent Fgf15 overexpression enhances cell cycle progression directly without liver surgery, levels of proteins involved in cell cycle progression were determined in Tg mice without (basal level) and with CA feeding. Without CA feeding, although the BA pool size was very low in the Tg mice, the basal mRNA levels of Ccnd1 (Cyclin D1), Pcna (proliferating cell nuclear antigeph [PCNA]), and Cdkn1a (cyclin-dependent kinase inhibitor 1A) (p21), but not Ccne1 (Cyclin E1), were significantly higher than those in WT mice (Fig. 2D). With CA feeding, WT mice did not show an increase in the mRNA levels of Ccnd1, Ccne1, and Pcna except for Cdkn1a, which were highly induced. In Tg mice, CA further induced the expression of all these mediators (Fig. 2D). The protein expression of cell-signaling mediators related to cell proliferation and Fgfr4 was further determined. JNK and ERK signaling pathways were activated in the Tg mice, as evidenced by increased p-JNK and p-ERK levels, as well as their downstream targets including c-Jun and c-Fos, but not c-myelocytomatosis (c-Myc). CA increased the protein levels of Cyclin D1 and PCNA but reduced those of Cyclin E1 and c-Myc, in WT and Tg mice (Fig. 2E). Overall, these results suggest that Fgf15 may directly promote cell proliferation in addition to modulating BA levels at basal levels.
OVEREXPRESSION OF Fgf15 PROMOTES LIVER REGENERATION FOLLOWING 2/3 PHx
The induction of pro-proliferative genes in the liver by Fgf15 transgene prompted us to determine the effects of Fgf15 overexpression on liver regeneration at low BA levels. PHx was performed in Tg mice with overexpression of Fgf15 in both liver and intestine (to mimic human expression pattern)-Veh/PHx, and in Tg mice with Fgf15 overexpression only in the intestine by treatment with Dox for 5 days followed by Dox-free for 2 days (mouse expression pattern)-Dox/PHx (Fig. 3A). Twenty-four hours following PHx, the total BA pool size in Tg mice remained lower (Fig. 3B). Ccnd1 and Ccne1 gene expression started to increase 24 hours post PHx,22 and the current study showed that overexpression of Fgf15 in both liver and intestine induced Ccnd1, Ccne1, and Cdkn1a following PHx, whereas overexpression of Fgf15 only in the intestine induced Ccnd1 and Ccne1 to a lesser extent, and significantly increased levels of Cdkn1a (Fig. 3C), suggesting that Fgf15 in both liver and intestine contributes to cell proliferation after PHx. The ERK, but not JNK, pathway showed higher activation following PHx in Tg mice (Fig. 3D). The protein levels of Cyclin D1, Cyclin E1, Cdk4, PCNA, and c-Myc, but not c-Jun, were markedly increased in Fgf15 Tg mice (Fig. 3D).
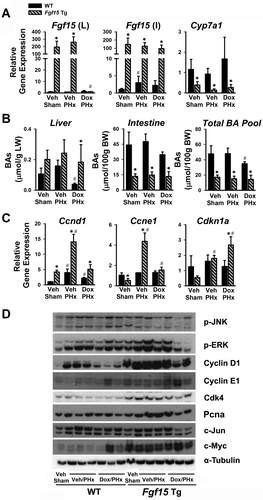
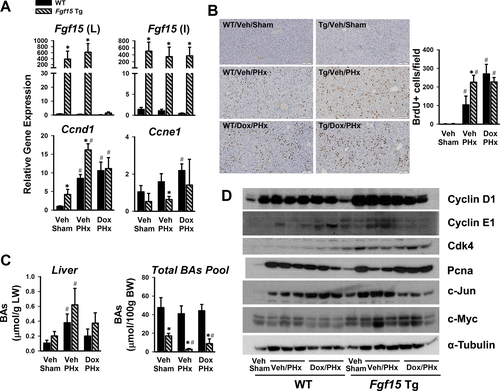
Thirty-six hours following PHx, overexpression of Fgf15 in the liver remained to be suppressed by Dox, but it was fully recovered in the intestine (Fig. 4A). At this time point, Ccnd1 gene expression was still much higher in mice with Fgf15 overexpression in both the liver and intestine (Veh/PHx), but not in Tg mice with overexpression of Fgf15 in intestine only (Dox/PHx). Ccne1 expression tended to be lower in Fgf15 Tg than WT mice, which may reflect a compensation. DNA proliferation, measured by BrdU incorporation in hepatocytes during liver regeneration, was higher in Tg mice with Fgf15 overexpression in both liver and intestine (Veh/PHx) compared with WT (Fig. 4B). Interestingly, Dox treatment markedly increased BrdU incorporation in WT mice with PHx (Dox/PHx), and to a similar degree when Tg mice was under the same treatment (Fig. 4B). At 36 hours after PHx, the BA pool size remained lower in Tg mice, but the hepatic BA levels were similar between WT and Tg mice, and it is noted that Tg mice tended to have higher BA levels in the liver (Fig. 4C). Fgf15 overexpression, regardless in both liver and intestine (Veh/PHx) or in intestine only (Dox/PHx), led to higher expression of cell cycle–related proteins, including Cyclin D1, Cdk4, and c-Myc (Fig. 4D). Intestine-overexpression of Fgf15 reduced c-Jun protein levels at this time point after PHx (Fig. 4D).
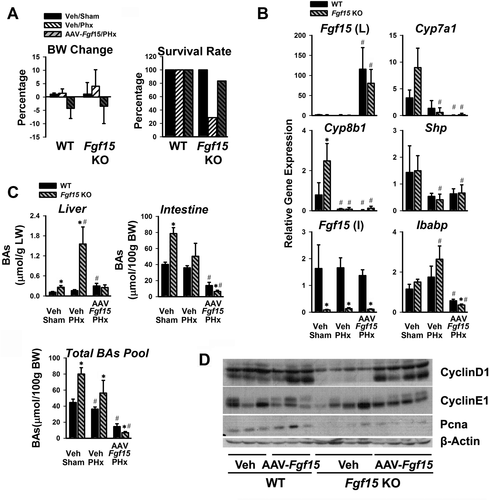
In another independent gain-of-function model, Fgf15 was overexpressed in the liver by AAV-mediated transduction in Fgf15 KO mice. Transduction by AAV-Fgf15 led to reduced BW, and decreased mortality following PHx (from 80% to 30%) (Fig. 5A). This liver-specific AAV-mediated Fgf15 overexpression reduced mRNA levels of hepatic Shp and intestinal Ibabp genes induced by BAs (Fig. 5B). The Fgf15 KO mice normally had higher BA levels (Fig. 5C), and following PHx, they presented with 8-fold to 10-fold higher hepatic BA concentration than WT mice (Fig. 5C). AAV-Fgf15 overexpression inhibited BA synthesis, resulting in a significant reduction in liver BA concentration, with the total BA pool size returning to levels in WT mice (Fig. 5C, AAV-Fgf15/PHx). Furthermore, the protein levels of Cyclin D1, Cyclin E1, and PCNA were induced in the AAV-Fgf15 mice (Fig. 5D).
Fgf15 ENHANCES HEPATOCYTE PRIMING IN LIVER REGENERATION FOLLWOING 2/3 PHx
The effects of Fgf15 on the priming of liver regeneration were examined at 30 minutes and 60 minutes following PHx in WT and Fgf15 Tg mice, by determining the mRNA and protein levels of genes involved in the initial stage of cell proliferation (Fig. 6). At 30 minutes, PHx increased the mRNA levels of c-Jun and c-Fos by 5-fold and 100-fold, respectively, in WT mice, and 13-fold and 250-fold, respectively, in Tg mice (Fig. 6A). This marked induction returned to baseline levels at 60 minutes (Fig. 6A). A marked induction of c-Myc and Igfbp1 was also observed, which reached their maximum expression at 60 minutes, and with significant difference in c-Myc expression between WT and Fgf15 Tg mice (Fig. 6A).
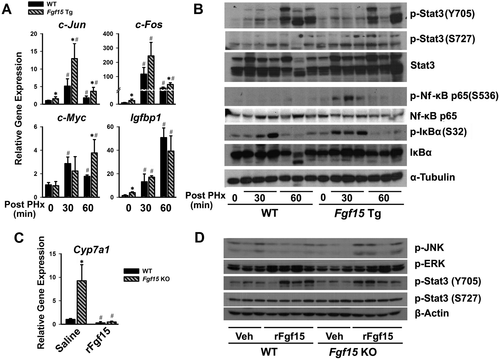
Activation of signal transducer and activator of transcription 3 (Stat3) with PHx, measured by increased p-Stat3 (S727) and p-Stat3 (Y705), is well-known as one of the major priming events during liver regeneration. The basal levels of p-Stat3 (S727) and p-Stat3 (Y705) appeared to be higher in Fgf15 Tg mice (0 minutes) (Fig. 6B). Both WT and Fgf15 Tg mice demonstrated increased activation of Stat3 at 30 minutes and 60 minutes after PHx, but Tg mice had a higher induction of p-Stat3 (S727) at 30 minutes (Fig. 6B). NF-κB signaling pathway is activated by IKK-mediated phosphorylation of the inhibitory subunit IκBα at Ser32 and Ser36, which leads to proteasomal degradation of inhibitor IκB, thus releasing NF-κB subunits and activating gene transcription in the nucleus.28 The phosphorylation and activation of NF-κB (p65) at Ser536 and p-IκBα at Ser32 in Fgf15 Tg mice was significantly higher than in WT mice at 30 minutes after PHx, and returned to basal levels in both strains at 60 minutes after PHx. There were no significant changes in protein level for NF-κB (p65) and IκBα. The basal levels of c-Jun, c-Fos, and c-Myc tended to be higher in Tg mice (0 minutes) but were not further induced by PHx in both strains (Supporting Fig. S2).
To further determine the signaling cascades activated by Fgf15, Fgf15 KO mice were injected with the recombinant Fgf15 protein (rFgf15) that was developed in our lab. The rFgf15 was functional, as it significantly suppressed Cyp7a1 gene expression (Fig. 6C). Despite having little effect on the gene expression of c-Jun and c-Fos (Supporting Fig. S3), injection of rFgf15 increased p-JNK, p-ERK and p-Stat3, indicating that Fgf15 could activate both mitogen-activated protein kinase (MAPK) and Stat3 signaling cascades in vivo (Fig. 6D).
Fgf15 MAY REGULATE THE TERMINATION OF LIVER REGENERATION AFTER 2/3 PHx
In mice, the liver regenerates and grows back to its original mass within 14 days after 2/3 PHx. Hepatocytes reenter the cell cycle and replicate after incision and return to quiescence state after liver recovers its mass.2 However, the mechanisms for the termination of regeneration are complex and not fully understood. Extensive studies have elucidated that the hippo pathway plays a critical role in organ size determination and is involved in the termination stage of liver regeneration. Activation of the hippo pathway leads to phosphorylation of Yes-associated protein (Yap) and transcriptional co-activator with PDZ binding motif (Taz), which promotes Yap/Taz subsequent cytoplasmic sequestration and inactivation.29, 30
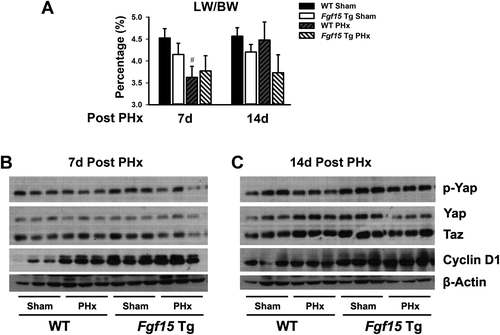
The effects of Fgf15 on liver regeneration termination is unknown. To study this effect, activation of the hippo pathway was assessed in Fgf15 Tg mice at day 7 and day 14 after PHx (Fig. 7). Consistent with the previous result, Fgf15 Tg mice showed lower LW/BW ratio compared with WT mice (Fig. 7A, Sham). Livers in WT mice were still regenerating at day 7 after PHx and recovered their original weight 14 days after, whereas livers in Fgf15 Tg mice stopped growing at day 7 following PHx and kept a similar weight at 14 days after PHx (Fig. 7A). Consistent with the LW changes, Cyclin D1 level was still much higher in WT mice 7 days following PHx, but similar in Fgf15 Tg mouse livers between Sham and PHx, indicating that hepatocyte replication remained active in WT but not in Tg mice (Fig. 7B). There were no significant differences for p-Yap, Yap, and Taz in WT mice between Sham and PHx, but in Fgf15 Tg mice, a slight decrease in p-Yap and Taz levels following PHx, indicating an activation of the hippo pathway in Fgf15 Tg mice 7 days after PHx (Fig. 7B). At day 14 after PHx, hepatocytes in both WT and Fgf15 Tg mice returned to a more quiescence state, revealed by similar Cyclin D1 levels between the Sham and PHx groups (Fig. 7C). Compared with the WT/Sham group, the protein levels of p-Yap, Yap, and Taz were much higher in Fgf15 Tg mice (Fig. 7C). PHx decreased p-Yap (inactive form) but increased Yap (active form) and Taz expression in WT. However, compared with Sham, PHx significantly decreased both p-Yap and Yap in Fgf15 Tg mice (Fig. 7C). These data indicate that after PHx, hepatocytes from WT and Fgf15 Tg mice may have entered termination stage differently and Fgf15 may have promoted the hippo pathway activity after hepatectomy in mice.
To determine the effects of Fgf15 on changes in biliary mass after PHx, biliary mass in WT and Fgf15 Tg mice was measured at 7 days and 14 days after PHx (Supporting Fig. S4). However, there was no significant difference in the biliary mass between WT and Fgf15 Tg mice.
Discussion
BAs are synthesized in the liver from cholesterol and primarily function to facilitate the absorption of lipids and fat-soluble vitamins in the intestine.3 Moreover, BAs are a component of bile, undergo enterohepatic circulation, and act as signaling molecules.3, 4, 10 An emerging function of BAs is to promote liver regeneration. However, accumulation of BAs is toxic and can lead to cellular damage and death, cholestasis, inflammation, and tumor genesis.3, 8, 9, 31 BA receptors, including FXR and TGR5, are critical for maintaining BA homeostasis, and deficiency of FXR and TGR5 in mice results in impaired liver regeneration after 2/3 PHx.16, 20 It has previously been suggested that BAs are involved in liver cell proliferation, and physiological concentrations of BAs may activate FXR to promote liver regeneration.16 Findings from our laboratory and other groups have identified that a FXR target gene in the intestine, Fgf15 (human orthologue, FGF19), is critical in promoting liver regeneration and maintaining liver size following PHx, but not after chemical-induced liver injury.19, 21, 22, 32 Consensus indicates that FGF15/19 suppresses BA synthesis and reduces total BA pool size,13, 14 but it is unclear to what degree Fgf15 directly stimulates cell proliferation in vivo. Therefore, through generation of the Fgf15 Tg mice overexpressing Fgf15 in liver and/or intestine by supplementing these mice with or without CA, we are able to use this model to distinguish the direct role of Fgf15 in promoting hepatocyte proliferation and liver regeneration in vivo versus the indirect effects mediated through BA suppression.
Human FGF19 is abundantly expressed in the gallbladder, intestine, and common bile duct, as well as in hepatocytes during cholestasis.33 Contrastingly, mouse Fgf15 is only expressed in the intestines. The Fgf15 Tg mice generated in the current study expressed Fgf15 in both the liver and intestine, with the transgene driven by the promoter of FABP1. There are two major benefits using this model system. First, mouse Fgf15 but not human FGF19 gene is overexpressed in these mice, as a previous publication has shown that FGF19 may not activate Fgfr4 in mice19; therefore, this model is suitable to study Fgf15 function in vivo. Second, by using the Tet-Off system, we overexpressed Fgf15 transgene expression in both the liver and intestine without Dox treatment; and with Dox treatment, rebound overexpression is only achieved in the intestine. This generates a more human relevant mouse model. Indeed, the results from current study showed that overexpression of Fgf15 in either liver plus intestine or in intestine only promotes hepatocyte proliferation and liver regeneration, suggesting that Fgf15 is mitogenic regardless of tissue origin.
FGF15/19 is secreted from the enterocytes under physiological conditions, circulates to the liver, and activates Fgfr4 in the presence of β-Klotho. Activation of Fgfr4 leads to activation of several signaling pathways, including MAPK and Stat3, which are believed to be the major mechanism to suppress BA synthesis and promote cell cycle progression.13, 22, 34 Activation of these pathways could contribute to the priming of liver regeneration. The early stage of liver regeneration (approximately 0 hours to 4 hours after PHx in mice) is known as the “priming” phase, which requires an orchestration of various molecular signaling pathways and complex tissue interactions. In this stage, a wide variety of genes is differentially expressed and cytokine signaling pathways are activated to prepare for the regeneration process.2, 35 Cytokines of Tnfα and Il-6,36, 37 and activation of signaling cascades of the transcription factors Nf-κB and Stat3, play critical roles in the priming phase and triggering of the G0/G1 transition of the cell cycle.28, 38, 39 Our current results suggest that activating Nf-κB is also an important mechanism for Fgf15-mediated priming in liver regeneration. Specifically, the Tg mice showed greater activation of NF-κB than Stat3 or ERK minutes following PHx. The mechanism of NF-κB activation needs to be further studied in the future, as contrasting results have been reported that Fgfr4 activation inhibits NF-κB signaling.40
The hippo pathway has been demonstrated to be a key regulator of organ size determination and tumor suppression in mammals.29, 30 Activation of the hippo pathway leads to Yap phosphorylation and thus decreased Yap nuclear localization,30 so Yap/Taz activation in hepatocytes induces cell senescence and suppresses transcription activation. Total Yap and Taz levels were higher in Fgf15 Tg mice, suggesting that the hippo pathway was more active in Fgf15 Tg mice (Fig. 7) and Fgf15 might be involved in the suppression of cell proliferation in the termination stage of liver regeneration.
Mst1/2 and Sav1 are upstream core components of hippo signaling, and activation of Mst1/2 and Sav1 leads to phosphorylation of Yap/Taz. However, Mst1 and Sav1 protein levels in Fgf15 Tg mice did not show any significant differences (data not shown), indicating that the phosphorylation of Yap/Taz might be independent of the activation of Mst1/2 and Sav1 pathways. Previous studies have shown that Yap/Taz can be phosphorylated by many other kinases, including JNK, EGFR-Ras-MAPK, and the Src family tyrosine kinases.41 It is well known that Fgf15/FGF19 activates JNK and ERK, but it is still unclear how this Yap/Taz suppression signaling cooperates with cell proliferation signaling to precisely regulate cell division and hepatocyte proliferation during liver regeneration.
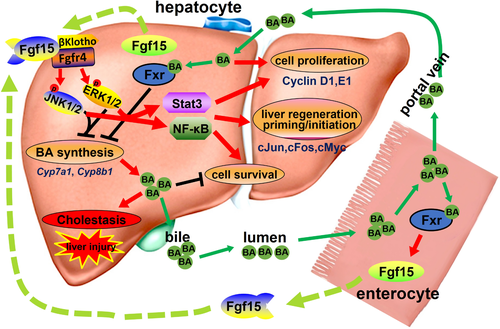
The most important finding of this study is that we demonstrate how Fgf15 directly stimulates cell proliferation under low BA levels, suggesting that Fgf15 could directly promote cell proliferation independent of BA suppression during liver regeneration. Our study is in agreement with previous reports showing that Fgf15 is responsible for the effects of bile salts on liver growth and the direct mitogenic activity of FGF19 on primary hepatocytes and cultured cholangiocytes in vitro.21, 33 This study provides evidence to show that Fgf15 directly promotes liver growth in vivo independent of BA levels. In summary, Fgf15 effectively suppresses BA synthesis regardless of tissue origin, and following PHx, Fgf15-mediated suppression of BA synthesis aids in the prevention of BA toxicity (Fig. 8). Furthermore, Fgf15 directly promotes cell proliferation and liver regeneration by activating signaling pathways that drive proliferation and cell survival (Fig. 8). In conclusion, we provide evidence showing that Fgf15 mediates an important crosstalk by which the intestines directly stimulate liver regeneration.
Potential conflict of interest
Nothing to report.
REFERENCES
Abbreviations
-
- AAV
-
- adeno-associated virus
-
- BA
-
- bile acid
-
- BrdU
-
- bromodeoxyuridine
-
- BW
-
- body weight
-
- CA
-
- cholic acid
-
- CAR
-
- constitutive androstane receptor
-
- Cdkn1a
-
- cyclin-dependent kinase inhibitor 1A
-
- c-MYC
-
- c-myelocytomatosis
-
- Dox
-
- doxycycline
-
- ERK
-
- extracellular signal-regulated kinase
-
- FABP
-
- fatty acid binding protein
-
- FGF15/19
-
- Fibroblast growth factor 15/19
-
- FXR
-
- farnesoid X receptor
-
- GPCR
-
- G protein-coupled receptor
-
- H&E
-
- hematoxylin and eosin
-
- JNK
-
- c-Jun N-terminal kinase
-
- KO
-
- knockout
-
- LW
-
- liver weight
-
- MAPK
-
- mitogen-activated protein kinase
-
- PCNA
-
- proliferating cell nuclear antigeph
-
- PHx
-
- partial hepatectomy
-
- rFgf15
-
- recombinant Fgf15 protein
-
- S1PR2
-
- sphingosine 1-phosphate receptor 2
-
- STAT3
-
- signal transducer and activator of transcription 3
-
- Taz
-
- transcriptional co-activator with PDZ binding motif
-
- TC
-
- total cholesterol
-
- Tg
-
- transgenic
-
- TG
-
- triglycerides
-
- TGR5
-
- G protein-coupled bile acid receptor 1
-
- TRE
-
- tTA-responsive element
-
- tTA
-
- tetracycline transactivator
-
- WT
-
- wild type
-
- Veh
-
- vehicle
-
- and Yap
-
- Yes-associated protein




