Nonstructural protein 5A/P32 deletion after failure of ledipasvir/sofosbuvir in hepatitis C virus genotype 1b infection
Potential conflict of interest: Dr. Kodama received grants from Gilead. Dr. Takehara is on the speakers' bureau for and received grants from Gilead.
This work was partially supported by a Grant-in-Aid for Research on Hepatitis from the Japan Agency for Medical Research.
Abbreviations
-
- DAAs
-
- direct-acting antivirals
-
- NS5A
-
- nonstructural protein 5A
-
- RASs
-
- resistance-associated substitutions
-
- P32del
-
- P32 deletion
-
- VF
-
- virological failure
Nonstructural protein 5A (NS5A) inhibitors play an essential role in the combination treatment of direct-acting antivirals (DAAs) against chronic hepatitis C (CHC). Daclatasvir, a prototype of this class, combined with asunaprevir, has been widely used since 2014 in Asia, where genotype 1b infection is prevalent. Well-known resistance-associated substitutions (RASs) for this treatment include NS5A-L31M/V and Y93H. In addition, recent studies have reported the emergence of a deletion mutant at NS5A-P32 in a subset of patients at the time of virological failure (VF) with this treatment.1 This mutant is attracting much attention because replicon studies indicate that, compared with NS5A-Y93H, it confers extremely higher resistance to NS5A inhibitors.2 Moreover, retreatment with DAAs in patients carrying this mutant after daclatasvir/asunaprevir treatment has been reported to be ineffective.3 Here, we report on a case of NS5A-P32 deletion (P32del) after ledipasvir/sofosbuvir treatment.
We treated 1,031 Japanese patients with genotype 1 CHC with ledipasvir/sofosbuvir, and 12 cases exhibited VF. Among these cases, sera from 10 patients available were analyzed for NS5A RAS by deep sequencing, as described1 (Table 1). All patients had genotype 1b infection and, except for case no. 1, exhibited the NS5A Y93H substitution at baseline or after VF. Case no. 1 showed P32del at the time of VF; the P32del was not detected at baseline (0.1% deep sequencing cutoff), but appeared 4 weeks after completion of 12 weeks of treatment and was continuously detected at a high frequency for 52 weeks (Fig. 1A). After the appearance of the P32del, nucleotide sequences of NS5A domain-I in a major clone were completely consistent at any point. In addition, known RASs to sofosbuvir, including S282T, were not detected by deep sequencing during the entire course. The patient was a 75-year-old treatment-naïve man with compensated cirrhosis with a treatment history for hepatocellular carcinoma.
| RAS at the Start of Ledipasvir/Sofosbuvir Treatment | Duration | RAS After Failure of Ledipasvir/Sofosbuvir Treatment | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | R30 | L31 | P32 | Q54 | Y93 | of Therapy | Effect | Point | R30 | L31 | P32 | Q54 | Y93 |
| 1 | – | M (99.7%) | – | – | – | 12W | Relapse | Post 4W | – | M (84.5%) V (15.5%) | del (84.5%) | – | – |
| 2 | – | F (3.8%) V (3.1%) | – | – | H (62.5%) | 12W | Relapse | Post 4W | – | F (5.4%) V (3.6%) | – | – | H (99.7%) |
| 3 | – | V (12.9%) | – | – | H (34.9%) | 12W | Relapse | Post 4W | – | V (60.9%) | – | – | H (99.7%) |
| 4 | – | V (62.3%) M (37.1%) | – | H (32.3%) | H (96.5%) | 12W | Relapse | Post 4W | – | V (81.3%) M (18.3%) | – | H (14.4%) | H (99.6%) |
| 5 | Q (99.6%) | – | – | – | H (99.8%) | 12W | Relapse | Post 12W | N.A. | N.A. | N.A. | N.A. | N.A. |
| 6 | – | F (1.2%) | – | H (75.5%) | H (63.0%) | 12W | Relapse | Post 12W | – | – | – | H (99.9%) | H (99.7%) |
| 7 | Q (99.4%) | – | – | H (1.2%) | H (62.6%) N (35.7%) S (1.4%) | 12W | Relapse | Post 12W | Q (99.8%) | – | – | H (0.4%) | H (99.8%) |
| 8 | – | V (2.8%) | – | H (96.5%) | H (3.3%) | 4W | Relapse | Post 12W | – | V (24.2%) I (12.7%) | – | H (61.2%) | H (33.2%) |
| 9 | Q (94.3%) | – | – | – | H (6.4%) | 8W | Relapse | Post 12W | Q (96.5%) | – | – | – | H (99.8%) |
| 10 | – | – | – | – | – | 4W | Relapse | Post 12W | – | M (1.5%) | – | – | H (93.8%) N (5.3%) |
- A cut-off threshold of 0.1% was set. –, not detected; del, deletion. Point refers to point of resistance test after re-elevation of the hepatitis C virus HCV RNA viral load.
- Abbreviations: W, weeks; N.A., not available because of lack of success in generating PCR amplicons; del, deletion.
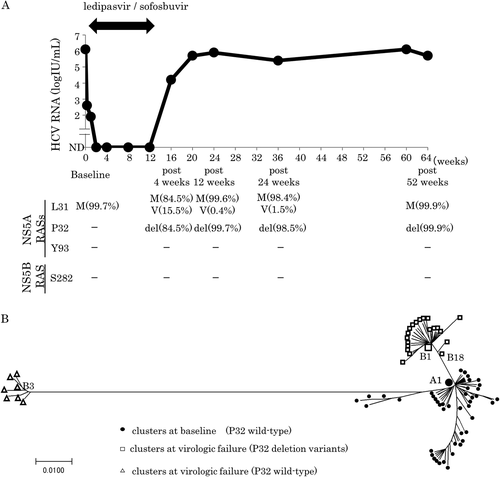
We performed phylogenetic tree analysis1 to clarify the relationship between pretreatment virus and posttreatment virus (Fig. 1B). This analysis indicated that the most frequent wild-type virus at baseline (A1 in Fig. 1B) acquired a deletion of three bases at position P32, becoming virus B18 posttreatment; one more sense mutation resulted in the most frequent virus, B1, posttreatment, which served as the starting point for the subsequent expansion of diversity.
Both clinical trials and real-world data have shown that the viral clearance rate of ledipasvir/sofosbuvir is over 95%. Recently, Wiles et al.4 analyzed posttreatment resistance in 51 patients (42, genotype 1a; 9, genotype 1b) who participated in ledipasvir/sofosbuvir phase I/II clinical trials and reported that the common substitutions were Q30R/H and/or Y93H/N for genotype 1a and Y93H for genotype 1b. However, these investigators did not report any instances of P32del. In our cohort, 1 of 10 patients with genotype 1b infection developed P32del, indicating that emergence of this deletion is not a specific event associated with daclatasvir/asunaprevir treatment, but can occur with other treatment regimens, at least in patients with genotype 1b infection. In our patient, the frequency of P32del variants increased after the time of VF and remained very high for an extended period of time, suggesting that the replication fitness of this variant is strong and therefore that its emergence may have an impact on the therapeutic effects of DAA retreatment using NS5A inhibitors. We should pay more attention to the appearance of this rare variant in DAA failure and may need to consider what type of retreatment is appropriate for this specific, refractory variant. Considering the extremely high resistance of this variant to NS5A inhibitors, DAA regimens containing both an NS3/4A inhibitor and sofosbuvir, addition of ribavirin or longer-duration therapies might be treatment options for patients with P32del.




