A2AR-induced transcriptional deregulation in astrocytes: An in vitro study
Funding information: ANR (GRAND, SPREADTAU, ADORATAU & ADORASTrAU); DFG Center for Nanoscale Microscopy and Molecular Physiology of the Brain (CNMPB); Fondation pour la Recherche Médicale, Vaincre Alzheimer, Fondation Plan Alzheimer; Inserm, CNRS, Université Lille, Lille Métropole Communauté Urbaine, Région Hauts-de-France, DN2M; programs d'investissements d'avenir LabEx (excellence laboratory) DISTALZ (Development of Innovative Strategies for a Transdisciplinary approach to ALZheimer's disease); SFB1286 project B6
Abstract
Adenosine A2A receptors (A2AR) are modulators of various physiological processes essential for brain homeostasis and fine synaptic tuning. In certain neurodegenerative conditions, notably Alzheimer's disease (AD), A2ARs are pathologically upregulated in neurons but also in astrocytes. In that context, the use of A2ARs inhibitors, normalizing impaired receptor function, is seen as a potential therapeutic strategy. However, the impact of A2AR alterations, particularly in astrocytes, is not fully understood. Here, we investigated the effect of A2AR overexpression on transcriptional deregulation in primary astrocytic cultures. By performing whole transcriptome analysis, we found that A2AR overexpression promotes robust transcriptional changes, mostly affecting immune response, angiogenesis, and cell activation-related genes. Importantly, we observed that treatment with SCH58261, a selective A2AR antagonist, restored the expression levels of several inflammatory and astrocytic activation-related genes, such as Interleukin-1beta and vimentin. This supports the notion that A2AR blockade could restore some astrocytic dysfunctions associated with abnormal A2AR expression, further arguing for a potential beneficial impact of receptor antagonists in A2AR-induced transcriptional deregulation, inflammation, and astrogliosis. Overall, our findings provide novel insights into the putative impact of A2AR overexpression on transcriptional deregulation in astrocytes, thereby opening novel avenues for the use of A2AR antagonists as potential therapeutic strategy in neurodegenerative diseases.
1 INTRODUCTION
Astrocytes are the most abundant glial cells in the brain and play a central role in brain homeostasis, as they modulate synaptic activity and plasticity, provide metabolic and structural support to neurons, and regulate ion and neurotransmitter homeostasis (Sofroniew & Vinters, 2010; Verkhratsky & Nedergaard, 2018). Structural and functional deregulation of astrocytes is known to be associated with neuropathological conditions, including neurodegenerative diseases, such as Alzheimer's disease (AD) (González-Reyes et al., 2017; Verkhratsky et al., 2010). It was previously shown that activation of astrocytes, in a state they undergo major transcriptional, morphological, and functional changes leading to impaired brain homeostatic function, is strongly associated with neurological diseases (Furman et al., 2012; Pekny et al., 2016; Liddelow et al., 2017).
Adenosine signaling plays a complex role in a number of physiological and pathophysiological processes (Cunha, 2016). The effects of adenosine in the brain are essentially mediated through the activation of A1 and A2A receptors. In physiological conditions, A2ARs play an important role in fine synaptic tuning (Cunha, 2016): neuronal A2ARs regulated NMDA receptors to control synaptic plasticity processes (Rebola et al., 2008; Temido-Ferreira et al., 2018) while astrocytic A2ARs control glutamate and GABA uptakes (Lopes et al., 2002; Matos et al., 2012, 2013; Cristovao-Ferreira et al., 2013), regulating the excitatory/inhibitory balance. Interestingly, reactive astrocytes have been shown to exhibit adenosine A2AR upregulation in different neuropathological conditions such as AD, epilepsy, Sandhoff disease, and also in animal models of AD and PD (Orr et al., 2015, 2018; Barros-Barbosa et al., 2016; Zhao et al., 2017; Faivre et al., 2018; Lee et al., 2018; Yu et al., 2008; Hu et al., 2016; Ogawa et al., 2018). However, the pathophysiological impact of such astrocytic A2AR upsurge remains unclear. In the present study, we provide data showing that the upregulation of A2ARs in primary cortical astrocytes strongly modulates astrocytic transcriptome with a significant impact on inflammatory, angiogenesis, and cell activation-related genes. Importantly, we confirmed that some of the changes were reversed by a selective A2AR antagonist. The present study provides novel insights into the consequences of astrocytic A2AR upregulation and supports the use of selective antagonists as a promising therapeutic strategy for several neurodegenerative diseases.
2 MATERIALS AND METHODS
2.1 Primary astrocyte cell culture
Primary astrocyte cultures were prepared from cerebral cortices of P1–P4 mouse pups. After decapitation, brains were kept in ice cold Hanks' Balanced Salt Solution (HBSS) (Ca2+ and Mg2+ free), cortex was isolated and meninges were removed. Tissue was digested by adding a solution of 0.05% Trypsin in HBSS (at 37°C for 15 min). Cells were incubated for 2–3 min at room temperature (RT) in Dulbecco's modified eagle medium (DMEM) containing 1% Gentamycin, 1% fungizone, and 10% fetal bovine serum, followed by trituration up to obtain a cell homogenate subsequently centrifuged at 800 rpm for 2 min. After discarding supernatant, the pellet was resuspended in DMEM medium containing 1% Gentamycin, 1% fungizone, and 10% fetal bovine serum. Cells from three brains were plated in a 175 cm2 flask in a DMEM medium volume of 50 mL. Mixed culture of astrocytes was incubated at 37°C with 5% CO2 for 3 days. At 3 days in vitro (DIV) medium was changed. At DIV 10 the mixed culture was shacked at 320 rpm overnight at 37°C. The remaining cells attached to the flask were washed with warm phosphate buffer saline (PBS) 1× and cells were further detached by a mild trypsinization procedure using PBS 1× with 0.25% trypsin at 37°C for 3–5 min. Culture medium was added to arrest trypsin action, and the cell suspension was collected and centrifuged at 1,000 rpm for 5 min. Cell pellet was resuspended in medium and cell count was performed. Finally, cells were seeded again with fresh medium on poly-l-lysine-coated plates or coverslips (for immunocytochemistry), at 1 × 106 or 0.05 × 106 cells/mL density. Astrocytes were further cultured for 2 days (48 hr) before the experiments were conducted.
In order to generate A2AR-overexpressing cells (A2AR-OE), primary astrocytic cultures were infected at DIV 7 with equimolar amounts of lentiviruses for either murine A2AR or Green Fluorescent Protein (GFP) as a control. The selective A2AR antagonist SCH 58261 (Tocris Bioscience) was applied to the primary astrocyte cultures at DIV 12 at a final concentration of 100 nM and cells were collected after 24 hr of treatment (DIV 13).
2.2 RNA-sequencing
Total RNA was extracted 6 days after infection and 24 hr after treatment with SCH 58261 using TRIzol Reagent (Thermo Fischer Scientific), with the protocol provided by the manufactures, followed by phenol-chloroform method. RNA-sequencing (RNA-seq) library preparation and sequencing were performed at the Transcriptome and Genome Analysis Laboratory, NGS-Core Unit of the University Medical Center Göttingen. Quality and integrity of RNA were assessed with the Fragment Analyzer from Advanced Analytical by using the standard sensitivity RNA Analysis Kit (DNF-471) (Agilent). All samples selected for sequencing exhibited an RNA integrity number over 8. RNA-seq libraries were performed using 500 ng total RNA of a non-stranded RNA-Seq, massively parallel mRNA sequencing approach from Illumina (TruSeq RNA Library Preparation Kit v2, Set A; 48 samples, 12 indexes, Cat. No. RS-122-2001). Specifically, we first optimized the ligation step diluting the adapters concentration to increase ligation efficiency (>94%), and finally we reduce the number of Polymerase Chain Reaction (PCR) cycles (10 cycles) to avoid PCR duplication artifacts as well as primer dimers in the final library product. Libraries were prepared on the automation (Beckman Coulter's Biomek FXP workstation). For accurate quantitation of cDNA libraries, a fluorometric based system, the QuantiFluor™dsDNA System, from Promega was used. The size of final cDNA libraries was determined by using the dsDNA 905 Reagent Kit (Fragment Analyzer from Advanced Bioanalytical) exhibiting a sizing of 300 bp on average. Libraries were pooled and sequenced on the Illumina HiSeq 4000 (SE; 1 × 50 bp; 30–35 Mio reads/sample). RNA-seq was performed from four independent cultures (different litters) per group (GFP and A2AR overexpressing astrocytes). The data set generated in this study (RNA-seq) has been submitted to the Sequence Read Archive (SRA-BioProject ID: PRJNA548046).
2.3 Raw read and quality check of RNA-seq
Sequence images were transformed with Illumina software BaseCaller to BCL files, which was demultiplexed to fastq files with bcl2fastq v2.17.1.14. The quality check was done using FastQC (Andrews, Simon; "FastQC a quality-control tool for high-throughput sequence data," https://www.bioinformatics.babraham.ac.uk/projects/fastqc/(2014)) (version 0.11.5, Babraham Bioinformatics).
2.4 Differential expression analysis
Differential expression analysis was performed as previously described (Halder et al., 2016). Briefly, sequencing data were processed by using a customized in-house software pipeline. Illumina's bcl2fastq (v1.8.4) was used to convert the base calls in the per-cycle BCL files to the per-read FASTQ format from raw images. Along with base calling, adapter trimming and demultiplexing were performed. To assess the quality control of the raw sequencing data, FastQC (v 0.11.5) was used. Reads were mapped to the mouse genome (Mus_musculus.GRCm38.86) using rna-STAR (version STAR_2.5.2b) (Dobin & Gingeras, 2015) in the non-splice-junction-aware mode and all other parameters were set in default settings by rna-STAR. In order to identify differentially expressed genes, we first identified unwanted sources of variation and removed them by using RUVSeq tool (Remove Unwanted Variations, RUVs, v. 1.8.0). We then used DESeq2 (version 1.14.1) (Love et al., 2014) to perform the differential expression analysis.
2.5 Quantitative real-time PCR
Total RNA was extracted from cells and purified using the NucleoSpin RNA Kit (Macherey-Nagel). One microgram of total RNA was reverse-transcribed using the HighCapacity cDNA reverse transcription kit (Applied Biosystems). qPCR analysis was performed on an Applied Biosystems StepOnePlus Real-Time PCR Systems using Power SYBRGreen PCR Master Mix (Applied Biosystems). The thermal cycler conditions were as following: 95°C for 10 min, then 40 cycles at 95°C for 15 s and 60°C for 25 s. Sequences of primers used in this study are given in Table S1. Cyclophilin A (Ppia) was used as a reference housekeeping gene for normalization. Amplifications were carried out in duplicate and the relative expression of target genes was determined by the ΔΔCt method.
2.6 Immunoblotting
For protein quantification, astrocytes were lysed with RIPA buffer [0.1% Triton X-100, 0.15M NaCl, 50 mM Tris pH 7.5, and a protease inhibitor cocktail tablet (Roche)]. The lysates were assayed for protein content with the Bradford assay (Bio-Rad) in an Infinite M200 PRO plate reader (Tecan Lta). The samples were mixed with 5× Laemmli buffer (250 mM Tris pH 6.8, 10% sodium dodecyl sulfate, 1.25% Bromophenol Blue, 5% β-mercaptoethanol, 50% glycerol), heat for 5 min at 95°C, and loaded on 11% sodium dodecyl sulfate polyacrylamide (SDS-PAGE) gels. After electrophoresis, proteins were transferred to nitrocellulose membranes in a semi-dry system (Tans-Blot TurboTM blotting system). About 5% skim milk, prepared with 1× Tris base solution/0.05% Tween (TBST), was used for blocking for 1 hr at RT. Incubation with the primary antibodies, namely anti-GFAP 1:1,000 (Santa Cruz, sc33673, Mouse), anti-A2AR 1:1,000 (Fronter Institute, Af1000, Guinea Pig), anti-SCG2 (Secretogranin II/Chromogranin C) 1:1,000 (kindly provided by Maité Montero, Université de Rouen, Normandie, France, Rabbit), and anti-β-actin 1:1,000 (Sigma, Mouse), all diluted in TBST with 5% skim milk, was carried out overnight at 4°C. After, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies 1:10,000 (anti-mouse or anti-rabbit, GE Healthcare), for 2 hr at RT. Between steps, membranes were washed three times with TBST, for 5 min at RT. The membranes were revealed using Immobilon Western Chemiluminescent HRP Substrate (Millipore Corporation) and the chemiluminescent signals were detected using Fusion FX (Vilber Lourmat). Quantification of bands was performed using Image J software.
2.7 Immunocytochemistry
Cells on coverslips were fixed using 4% paraformaldehyde at RT, for 15 min. 1× PBS was used to wash the cells and then permeabilization was performed using 0.1% Triton/PBS, for 15 min at RT. After blocking with 3% BSA/PBS at RT for 1 hr, cells were incubated overnight at 4°C with primary antibodies anti-A2AR 1:1,000 (Fronter Institute, Af1000, Guinea Pig), anti-GFAP 1:1,000 (DAKO, z0334, Rabbit), anti-MBP 1:1,000 (BioLegend, 808402, Mouse), anti-III β-tubulin (TUJ1) 1:1,000 (Covance, MMS-435P-250, Mouse), anti-NeuN 1:1,000 (Millipore, MAB377, Mouse), anti-O4 (clone 81) 1:1,000 (Millipore, MAB345, Mouse), anti-Iba1 1:1,000 (Abcam, ab5076, Goat), and anti-CD31 (PECAM-1) 1:1,000 (BD Pharmingen, 550274, Mouse) diluted in blocking solution. Cells were washed with 1× PBS and then incubated with secondary Alexa Fluor antibodies (anti-rabbit or anti-mouse, 488 or 555, Life Technologies), prepared at 1:1,000 in blocking buffer, for 2 hr at RT. Cells nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) and then the coverslips were mounted with Mowiol. Immunofluorescence images of Figure S1 were acquired using Olympus IX81-ZDC epifluorescence microscope (40× magnification objective) or using Leica DMI 6000B epifluorescence microscope (Leica), with 10× magnification. High-resolution images (Figure 1a–h) were acquired using the Zeiss Airyscan feature, based on a LSM710 confocal microscope (Zeiss, Carl Zeiss-Strasse 22 73447 Oberkochen, Germany). The Airyscan can achieve a resolution of 140 nm along the x-y axis and 400 nm axially by utilizing light that otherwise is rejected by the confocal pinhole and increasing of the signal to noise ratio. It is an array of 32 sensitive gallium arsenide phosphide detector elements, arranged in a compound eye fashion.
2.8 Cytotoxicity assays
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium) assay and ToxiLight bioassay (Adenylate Kinase release-based assay) (Lonza) were used to assess cell metabolism and cell viability, respectively, accordingly to the manufacture recommendations. The microplate reader spectrophotometer (Infinite M200 fluorescence plate reader, Tecan, Hamburg, Germany) was used for measurements of both assays.
2.9 Statistical analysis
Data are presented as mean ± SEM of, at least, three independent experiments. Differences between mean values were determined using the Student's t test or one-way analysis of variance, followed by a post hoc Fisher's Least Significant Difference (LSD) test using GraphPad Prism Software. Unless otherwise mentioned, p-value <0.05 was considered to indicate a significant difference.
3 RESULTS
3.1 A2AR overexpression induces transcriptional deregulation in astrocytes
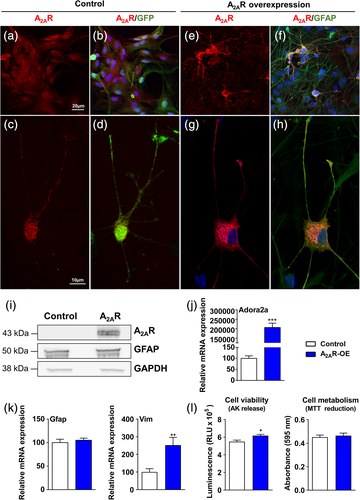
To investigate the impact of A2AR overexpression in astrocytes, we used primary murine astrocyte cultures infected with lentiviruses encoding either murine A2AR or GFP, as control. We first performed immunostainings in noninfected cells to assess the purity of the culture, and found a high percentage of astrocytes—79% of GFAP-positive cells, a standard astroglial marker; 14% of Iba1 positive cells, a microglial marker; no NeuN positive cells, and 2.6% of III β-tubulin positive cells, used as neuronal markers; 0.42% of Myelin basic protein (MBP) positive cells and no O4 positive cells were detected (oligodendrocyte markers); endothelial cells (CD31) were absent in the culture (Figure S1). Infected cells represented 72% of the total number of cells, suggesting we targeted mostly astrocytic cells. We confirmed A2AR overexpression using qPCR, immunoblotting analyses, and immunohistochemistry (Figure 1a–j). Using high magnification images, we were able to compare the cellular localization of A2AR in control astrocytes overexpressing GFP (Figure 1a–d) versus A2AR-overexpressing condition (Figure 1e–h). In the control cells, endogenous A2AR was predominantly localized at the cell membrane, with a typical membrane receptor puncta staining (Fig. 1c,d). On the other hand, astrocytes overexpressing A2AR displayed A2ARs both at the cell membrane as well as in the cytoplasm. (Figure 1g,h). At the morphological level, we did not observe any evident alterations upon overexpression of A2AR. Although A2AR overexpression in astrocytes had no impact on GFAP mRNA levels, the levels of Vimentin (Vim) were significantly increased (Figure 1k). Cell viability, assessed based on the release of adenylate kinase, showed a modest increase of toxicity in A2AR overexpressing astrocytes (Figure 1l). Cell metabolism, assayed using the MTT assay, showed no differences between A2AR overexpressing astrocytes and control cells (Figure 1l).
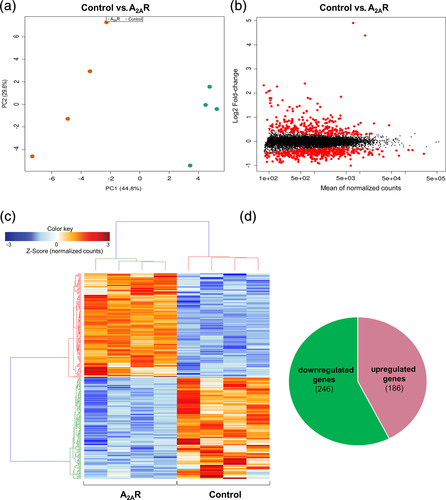
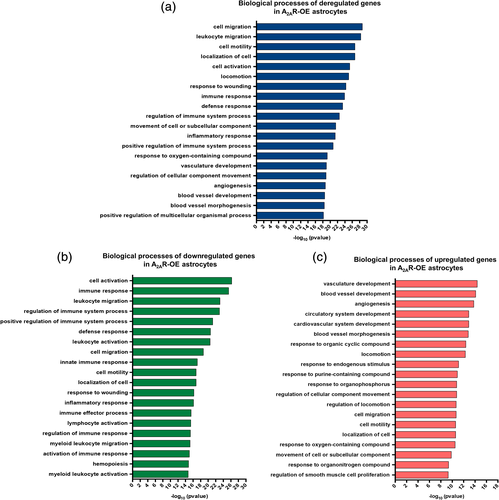
Next, to investigate the global changes induced by A2AR upregulation on the astrocytic transcriptome, we performed RNA-seq in primary astrocytic culture samples overexpressing either A2AR or GFP, as a control. Principal component analysis plots derived from RNA-seq data revealed a clear separation between control and A2AR expressing cultures (Figure 2a). Upregulation of the gene encoding for A2AR (Adora2a) was confirmed in RNA-seq data [log2Fold-Change(log2FC) = 4.90] for A2AR astrocytes versus control. MA plots, representing log ratio (M) and mean average (A), revealed a robust transcriptional deregulation associated with A2AR expression (Figure 2b). Heatmap plots also confirmed the differential regulation pattern of differentially expressed genes between A2AR-OE astrocytes and controls (Figure 2c). Considering a stringent p-adjusted value ≤0.01, we found 432 genes significantly modified (Table S2). Among those, 186 were upregulated and 246 downregulated. Differential analysis of count data also revealed the highest upregulation for Scg2, the gene encoding for Secretogranin II/Chromogranin C (log2FC = 4.39) followed by Il1ß (log2FC = 2.40). In contrast, Lipoprotein lipase showed the strongest downregulation (log2FC = −1.00). Interestingly, the Lipocalin-2 (Lcn2) gene, a pro-inflammatory gene previously associated with AD (Naudé et al., 2012), was found highly upregulated (log2FC = 1.56) (Table S2). To assess the molecular pathways affected by A2AR overexpression in astrocytes, we performed pathway analyses using ToppGene Suite. Using a cut off of padj ≤ 0.01 for significantly deregulated genes in A2AR expressing astrocytes, several pathways were identified as affected by A2AR overexpression. Interestingly, overall A2AR overexpression appears to deregulate genes mostly related to cell migration and activation, immune response, and angiogenesis (Figure 3a). In order to assess the effect of A2AR expression in overall upregulation and downregulation of the biological processes, both upregulated and downregulated genes were analyzed separately. In total, upregulated genes were involved in 695 pathways and the downregulated genes were related to 1,012 pathways (q value FDR < 0.05). The top 20 biological processes affected by A2AR expression revealed that pathways related to cell activation and immune response were mostly associated with downregulated genes (Figure 3b). On the other hand, angiogenesis-related processes were mainly related to upregulated genes found in A2AR-OE astrocytes (Figure 3c).
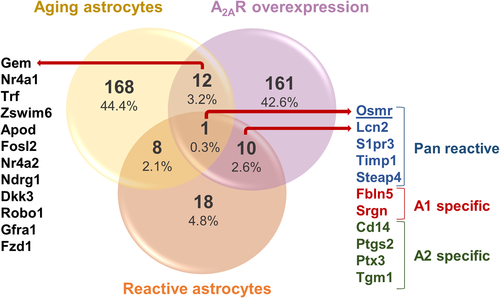
We also correlated our findings on A2AR-associated gene expression changes in astrocytes with changes associated with normal aging as well as neurodegenerative conditions (Figure 4), reactive astrocytes being strongly associated with neurodegenerative diseases, which are generally age-related disorders. For this, we used a publicly available data set of genes related to reactive astrocytes (Liddelow et al., 2017). We found that overexpression of A2AR in astrocytes induced an upregulation of 10 reactive astrocytic markers: five genes classified as pan reactive (Osmr, Lcn2, S1pr3, Timp1, and Steap4), two as A1 specific (neurotoxic; Fbln5, Srgn), and four as A2 specific reactive markers (neuroprotective; Cd14, Ptgs2, Ptx3, and Tgm1) (Zamanian et al., 2012) (Figure 4). A1-specific reactivation has been strongly linked to neurodegenerative processes (Liddelow et al., 2017). Furthermore, we retrieved the recent astrocyte transcriptomic data (cortex and hippocampus) associated with normal aging (Clarke et al., 2018), and found that 12 genes were commonly upregulated by A2AR and aging (Figure 4). Moreover, Osmr (a pan-reactive astrocytic marker) was found upregulated by A2AR expression in astrocytes, as well as by both astrocyte reactivation and aging process (Figure 4). We also analyzed a very recent data of single-cell transcriptomics from postmortem AD brains (Mathys et al., 2019) and we identified that the receptor tyrosine kinase, Mertk, and Cryl1 genes were downregulated in AD brains as well as in our A2AR overexpressing astrocytes (data not shown).
3.2 Overexpression of A2AR upregulates inflammatory and secretory-related genes in astrocytes
To further investigate the interaction between the genes affected by A2AR expression in astrocytes, we selected the top 50 deregulated genes (both upregulated and downregulated genes) determined by the strongest statistical significance (padj). Using the protein network STRING database, we observed a strong interaction between 23 out of 50 genes, including Adora2a (Figure 5a). Interestingly, this network was also formed by several top upregulated genes such as Il1ß, Lcn2, and Chitinase 3 Like 1 (Chi3l1), as mentioned above. These pro-inflammatory genes were previously described as linked to AD (Querol-Vilaseca et al., 2017; Shaftel et al., 2008; Naudé et al., 2012). Moreover, among the 27 other genes, the Solute Carrier Family 7 Member 11 (Slc7a11) and Scg2 were also found highly upregulated.
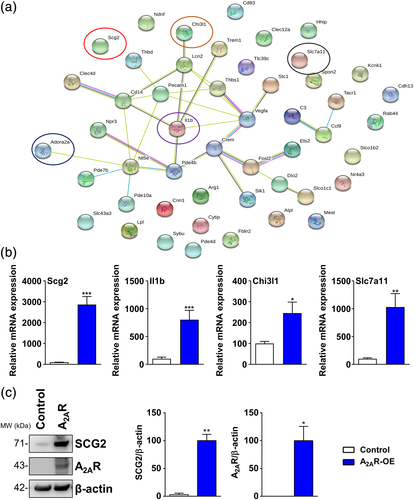
Next, in order to validate the RNA-seq data, we performed qPCR for Scg2, Il1ß, Chi3l1, and Slc7a11 genes in control and A2AR-OE astrocytes. As expected, we observed strong upregulation of all these genes associated with A2AR overexpression (Figure 5b). Since the neurosecretory protein Scg2 is deregulated in mouse models of AD (Saura et al., 2015) and is a predicted progression marker of the disease (Spellman et al., 2015), we further investigated its levels in A2AR-OE astrocytes. Importantly, we verified that Scg2 was highly increased upon A2AR overexpression in astrocytes (Figure 5c).
3.3 SCH 58261, a selective adenosine A2AR antagonist, rescues the expression of inflammatory and astrocyte activation genes
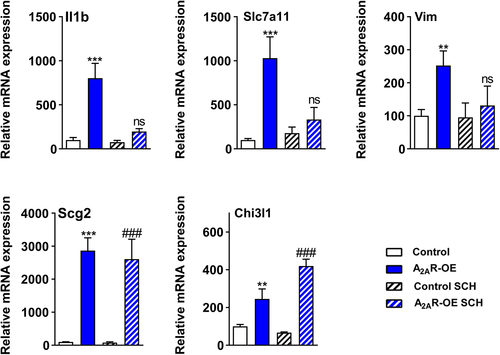
Given the strong effect of A2AR overexpression, we then asked whether the selective adenosine A2AR antagonist SCH 58261 could rescue the expression levels of several highly deregulated genes found in A2AR-OE astrocytes. In particular, we observed that the expression of the pro-inflammatory Il1ß and the cystine-glutamate antiporter, Slc7a11, which was found upregulated in A2AR-OE astrocytes, was restored upon SCH 58261 treatment. Moreover, treatment with SCH 58261 rescued the upregulation of Vim induced by A2AR expression in astrocytes. In contrast, SCH 58261 treatment did not change the upregulation of Scg2 and Chi3l1 induced by A2AR expression in astrocytes (Figure 6). Overall, SCH 58261 rescued the effect of A2AR-induced transcription deregulation of genes related to inflammation, cystine-glutamate antiport, and astrocytic reactivation.
4 DISCUSSION
Here, we provide new insights into the potential impact of A2AR upregulation on transcriptional deregulation in astrocytes. A2AR is a modulator of several physiological responses essential to brain homeostasis such as blood brain barrier maintenance, neurotransmitter release, and inflammation (Gołembiowska et al., 2013; Ouyang et al., 2013; Bhamidipati et al., 2013; Kim & Bynoe, 2015), processes that are highly associated with neurodegenerative diseases, such as AD and PD (Cunha, 2016). The relationship between A2AR and AD in the control of cognitive impairment has been extensively observed in several experimental models (Canas et al., 2009; Laurent et al., 2014; Laurent et al., 2014; Lee et al., 2018; Silva et al., 2018). Upregulation of A2AR in neurons was shown to induce aberrant phenotypic alterations during aging and in the early stages of AD (Temido-Ferreira et al., 2018). Early A2AR downregulation in neurons is sufficient to abrogate alterations of synaptic plasticity and memory in an animal model of amyloidogenesis (Viana da Silva et al., 2016). Neuronal A2AR dysregulation seems thus to play a crucial role in synaptic alterations underlying aging and AD. The astrocytic dysregulation might occur later on, presumably reinforcing the already ongoing synaptic alterations. However, the role of A2AR in glial cells is not well understood. Previous studies emphasized that astrocytic A2AR might control cognition in physiological and pathological situations (Barros-Barbosa et al., 2016; Orr et al., 2015; Matos et al., 2015). Astrocytic A2ARs were shown to control glutamate uptake through the control of Na+-K+-ATPase (Matos et al., 2012; Matos et al., 2013), gliotransmitter release (Cervetto et al., 2017), proliferation via Nfk-B (Ke et al., 2009), and the production of nitric oxide (Brodie et al., 1998). In the present study, we report, for the first time, the overall effect of A2AR overexpression—a process previously associated with various neurodegenerative conditions (Villar-Menendez et al., 2014; Laurent et al., 2014; Laurent et al., 2016; Orr et al., 2015; Albasanz et al., 2006)—on transcriptional deregulation, specifically in astrocytes. To do so, we used primary astrocytic cultures derived from mouse pups overexpressing A2ARs by lentiviral infection. In this model, we observed a 30-fold increase in A2AR (RNA-seq data). Several studies have shown that the levels of A2AR expression change in AD is up to five times higher than those of normal levels (Orr et al., 2015). Also, in vivo data from traumatic brain injury or amyloid-based models have demonstrated a similar range of A2AR upsurge ranging from 1.5 to 12 fold (Zhao et al., 2017; Orr et al., 2018; Faivre et al., 2018; Lee et al., 2018). One could therefore argue that the A2AR overexpression reached in our study is beyond the changes found in pathophysiological conditions. However, it is crucial to mention that in those studies the authors used whole tissue and not sorted cells suggesting that the actual level of dysregulation is certainly higher, probably reaching our experimental conditions. This will have to be solved in the future using fluorescence-activated cell sorting of astrocytes in vivo.
Overall, we found that A2AR overexpression has a greater impact on repressing gene expression, as most of the affected genes were downregulated. However, the top deregulated genes showing the highest fold-change, Scg2 and Il1ß, were upregulated. Interestingly, previous studies in rodents showed that Scg2 transcript and protein are abundant in cortical astrocytes and are increased in reactive astrocytes following brain injury (Paco et al., 2010). Our results revealed a strong upregulation of Scg2 in A2AR-OE astrocytes, at both mRNA and protein levels, suggesting that the A2AR potentially modulates the astrocytic secretory pathway, which might have an impact on glia–neuron interaction. Interestingly, the levels of Scg2 in human cerebrospinal fluid were previously suggested as putative marker of synaptic loss as well as of progression from mild cognitive impairment to AD (Spellman et al., 2015). However, it is noteworthy, that Scg2 is mainly a neuronal protein. In a context where neuronal A2ARs are early upregulated in AD brain (Viana da Silva et al., 2016; Temido-Ferreira et al., 2018), our data open the possibility that A2AR dysregulation might also involve early Scg2 neuronal alterations in AD brains, that will deserve further investigations.
Ontology analysis from our RNA-seq data showed that astrocytic A2AR deregulates mainly genes related to cell migration and activation, as well as immune response and angiogenesis. It has been suggested that A2ARs expressed by microglia, the main resident inflammatory brain cell, play a critical role in the control of neuroinflammation (Rebola et al., 2011; Boia et al., 2017). Some findings suggest that microglia A2ARs might also be dysregulated in the pathological context of AD (Orr et al., 2009; Angulo et al., 2003). Therefore, even the role of microglial A2AR remains to be uncovered in AD, our data support that astrocytic A2AR might also be relevant in neuroinflammatory processes underlying AD.
Despite most of the genes related to immune system were found downregulated, we found Il1ß, Lcn2, and Chi3l1, relevant pro-inflammatory genes, strongly upregulated upon A2AR overexpression in astrocytes. Notably, Lcn2 was previously associated with AD pathology as it was found increased in AD postmortem brain tissue (Naudé et al., 2012). Also, we found that angiogenesis-related genes were upregulated, according with a possible role of A2AR expression in inflammation. Consistently, it was previously suggested that A2AR activation in glia drives neuroinflammation and, consequently, the selective blockade of A2AR may be neuroprotective (Gyoneva et al., 2014; Minghetti et al., 2007; Nobre Jr. et al., 2010; Ogawa et al., 2018). Among many inflammatory cytokines linked to AD, IL1ß has been strongly involved in pathogenesis (Kitazawa et al., 2011; Sheng et al., 2000; Nicolia et al., 2017). In AD, IL1ß is essentially produced by microglial cells following maturation of pro-IL1ß by NLRP3 inflammasome (Heneka et al., 2018). Interestingly, adenosine has been shown to activate the NLRP3 inflammasome particularly through A2ARs in a macrophage cell line (Baron et al., 2015). It would be therefore tempting to speculate that the increase of IL1ß in A2AR-OE astrocytes might also involve activation of astrocytic NLRP3, known to be expressed by these glial cells (Heneka et al., 2018). In line with this, we observed that treatment with the A2AR antagonist SCH 58261 rescues the changes in Il1ß levels. The benefit afforded by inhibition of the NLRP3 or IL1ß signaling cascades in AD mouse models (Kitazawa et al., 2011; Heneka et al., 2013; Venegas et al., 2017) would be therefore prone to block a detrimental A2AR-mediated effect on IL1ß production by microglial and astrocytic cells. This hypothesis will need to be further investigated. Overall, these data raise the possibility that overexpression of A2AR in astrocytes, which is associated with neurodegeneration, particularly in AD, might increase neuroinflammation, and that this can be rescued by A2AR antagonists.
Furthermore, we found that Slc7a11, a gene encoding for a sodium-independent cystine-glutamate antiporter highly expressed in astrocytes, was upregulated in A2AR-OE astrocytes. In line with this, some studies suggest that the antiporter system Xc-, characterized by the import of the amino acid cystine into cells with a 1:1 counter-transport of glutamate, is upregulated in neuropathological conditions, particularly in AD patients (Bridges et al., 2012). Interestingly, the astrocytic system Xc- is responsible for the excitotoxic effects of Il1ß, which we found also upregulated in A2AR-OE astrocytes (Jackman et al., 2010; Fogal et al., 2007). Also, the Xc- deficiency was previously found to be protective in animal models of PD (Massie et al., 2011; Clark et al., 2010). Thus, we hypothesize that overexpression of A2AR in astrocytes might increase non-synaptic glutamate release via Xc- system, by upregulating Slc7a11, resulting in activation of ionotropic glutamate receptors and excitotoxic neuronal death (Lewerenz et al., 2013). Interestingly, we observed that treatment with SCH 58261 restored the levels of Slc7a11, suggesting a protective effect for the A2AR antagonist on excitotoxicity induced by high glutamate levels.
Given the known role of Vim and GFAP on astrocyte activation (Kamphuis et al., 2015), our data suggest that, although we did not observe changes on GFAP levels upon A2AR overexpression, Vim levels were significantly increased in A2AR-OE astrocytes. Thus, in accordance with a previous study using rat astrocytic cell line (Ke et al., 2009), our results indicate that A2AR overexpression in astrocytes might increase reactive astrogliosis, which is known to be an integral player on AD pathology (Osborn et al., 2016). Excitingly, 24 hr treatment with the A2AR antagonist, SCH 58261, was able to restore the expression levels of Vim, suggesting a protective effect of this antagonist on reactive astrogliosis. Interestingly, we found that overexpression of A2AR in astrocytes upregulates genes associated with reactive astrocytes as well as genes linked to normal aging process. This might indicate that, since reactive astrocytes are strongly linked to AD (Shi et al., 2017), the transcriptional changes induced by A2AR overexpression are linked to early activation of aging pathways as well as to neurodegeneration-related processes. The variety of reactive astrocytic types, such as pan reactive A1 (neurotoxic) or A2 (neuroprotective) specific (Zamanian et al., 2012), detected by RNA-seq might be due to the mixed population of astrocytes overexpressing A2AR and noninfected astrocytes. Thus, these reactivation mechanisms should be further explored. We also observed that the receptor tyrosine kinase Mertk was downregulated in A2AR astrocytes as well as in astrocytes of postmortem AD brains. MERTK pathway in astrocytes plays a critical role in synapse remodeling underlying neural circuit refinement, mediating synapse elimination, and has important implications in learning and memory as well in neurodegenerative diseases (Chung et al., 2013). Interestingly, this finding suggests that A2AR overexpression in astrocytes might interfere with mechanisms linked to synapse remodeling and, consequently, memory formation, both of which being impaired in AD.
The use of nonselective and selective A2AR antagonists, such as caffeine and SCH 58261, respectively, is considered as a potential therapeutic strategy for several neuropathological diseases (Boia et al., 2017; Laurent et al., 2014). In line with these studies, we suggest that this neuroprotective effect observed might be associated to a direct effect on astrocytic transcriptome by particularly downregulating the pro-inflammatory gene, Il1ß, as well as decreasing Vim expression. Noteworthy, we observed no normalization of Scg2 and Chi3l1 gene expressions upon treatment with SCH 58261, suggesting that a longer exposure to the treatment might be required to induce a stronger rescue effect on A2AR-induced transcriptional deregulations. A2AR are well known to form heteromers with several other G-coupled protein receptors: that is, A1 adenosine receptors, dopamine D2 receptors, CB1 cannabinoid receptors, mGluR5 metabotropic glutamate receptors, and presumably others (Ferre et al., 2018; Cunha, 2016 and references herein). All these receptors are notoriously expressed by astrocytes and sparse data suggest that A2A-A1 and A2A-D2 heteromers form in these glial cells, possibly controlling GABA and glutamate uptakes (Cristovao-Ferreira et al., 2013; Pelassa et al., 2019). Therefore, we cannot rule out that some of the transcriptional effects of A2AR astrocytic overexpression indirectly reflect modulation of the transduction of adenosine receptor's partners. This would eventually explain why some transcriptional effects of A2A upregulation are not reversed by the SCH 58261.
In conclusion, we provide novel insights into the role of A2AR dysregulation on astrocytic transcriptome, supporting that these receptors are promising targets in neurological disorders. Future in vivo studies will be required to confirm the extent of A2AR-mediated control in native brain astrocytes.
ACKNOWLEDGEMENTS
T.F.O. is supported by the DFG Center for Nanoscale Microscopy and Molecular Physiology of the Brain (CNMPB) and by SFB1286 project B6. The laboratory of L.B. and D.B. is supported by programs d'investissements d'avenir LabEx (excellence laboratory) DISTALZ (Development of Innovative Strategies for a Transdisciplinary approach to ALZheimer's disease), ANR (GRAND, SPREADTAU to L.B., ADORATAU & ADORASTrAU to D.B.), Région Hauts-de-France (COGNADORA), Fondation pour la Recherche Médicale, Vaincre Alzheimer, Fondation Plan Alzheimer (ADOEMOTAU) as well as Inserm, CNRS, Université Lille, Lille Métropole Communauté Urbaine, and DN2M. K.C. holds a doctoral grant from Lille 2 University. D.B. and T.F.O. received a funding from Campus France (MESRI/Deutscher Akademischer Austausch Dienst). The authors thank the Transcriptome and Genome Core Unit, University Medical Center Göttingen (UMG), G. Salinas and coworkers for their support with next generation sequencing. The authors would like to thank S. Trova, P. Giacobini (Inserm, UMRS1172), and the Bio Imaging Center Lille Nord-de-France Campus HU for their technical assistance on confocal microscopy.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.