Bromodomain-containing protein 2 gene polymorphism among patients with photosensitive epilepsy in Indonesia
Abstract
Objectives
Genetic-associated epilepsy in the Indonesian population is rarely discussed, and no study was specifically studied about photosensitive epilepsy. The fundamental goal of this research endeavor was to evaluate whether the single nucleotide polymorphism (SNP) of the Bromodomain-Containing Protein 2 (BRD2) gene gives vulnerability to photosensitive epilepsy among Indonesian descent.
Methods
This observational case–control study includes patients of Indonesian descent with Javanese ancestry. Clinical and neurophysiological data, along with electroencephalographic (EEG) recordings, were used to diagnose epilepsy and photosensitive epilepsy. Blood samples were collected and analyzed for BRD2 gene SNPs (rs206781, rs188245, and rs15912) using polymerase chain reaction (PCR), electrophoresis, and the Sanger sequencing method.
Results
This study included 27 participants, consisting of 17 patients in the epilepsy group (nine patients with photosensitive epilepsy and eight patients without photosensitive epilepsy) and 10 patients in the non-epilepsy group. Significant statistical differences were found in genotype (rs206781, p = 0.008 and rs188245, p = 0.004) and allele frequencies (rs206781, p < 0.001 and rs188245, p < 0.001) of the BRD2 gene in Indonesian descent with Javanese race patients diagnosed with photosensitive epilepsy and in those without this condition.
Significance
Our study corroborates the observation that genetic diversity within the BRD2 locus (rs206781 and rs188245) is associated with PE in Indonesian descendants of the Javanese race. To acquire a complete knowledge of the development of photosensitive epilepsy, further polymorphism studies at other SNP locations or genes are necessary.
Plain Language Summary
This study investigated whether genetic differences in the BRD2 gene were linked to photosensitive epilepsy (a type of epilepsy triggered by visual stimuli like flashing lights) in individuals of Indonesian Javanese descent. We analyzed deoxyribonucleic acid (DNA) samples from patients with epilepsy, including those with photosensitive epilepsy, and found that certain variations in the BRD2 gene were significantly more common in people with photosensitive epilepsy. These findings imply that genetic factors, specifically variations in the BRD2 gene, could elevate the risk of individuals in this population experiencing photosensitive epilepsy.
Key points
- This is the first genetic mutation study among photosensitive epilepsy patients in Indonesian populations.
- Seventeen patients in the epilepsy group (nine with photosensitive epilepsy) and 10 patients in the non-epilepsy group were included.
- Their DNAs were analyzed for the BRD2 gene (rs206781, rs188245, and rs15912) single nucleotide polymorphism.
- There was a significant association between the genotype and allele frequencies (rs206781 and rs188245) of the BRD2 gene in Indonesian descent.
- No significant difference was observed in the genotype and allele frequencies of rs15912.
1 INTRODUCTION
Reflex epilepsy is an epileptic seizure induced by a specific trigger, either by an external stimulus, an internal process, or activity.1 Photosensitive epilepsy (PE) constitutes a subtype of reflex epilepsy, characterized by seizures precipitated by visual stimuli, like flickering lights.1 It is the primary type of reflex epilepsy, significantly affecting 75%–80% of all cases. Common triggers include flashing lights from screens.2 Photosensitivity in epilepsy is prevalent among teenagers and females aged 7–19 years old.3 In photosensitive patients with intrinsic visual cortex hyperexcitability, seizures are induced by critical neuronal mass stimulation in the occipital cortex that may predispose them to large-scale neural activation.4 PE is also correlated with numerous types of seizures, that is, generalized tonic–clonic, generalized myoclonic, eyelid myoclonus, focal, and absence seizures.2
Recent previous research suggested a relationship between specific gene variations and PE. However, the exact mechanism is still unclear, and different studies have yielded varying results. A genetic region on chromosome 6p215, 6 has been identified as potentially influencing both electroencephalographic photo-paroxysmal response (PPR) and juvenile myoclonic epilepsy (JME). These conditions share neurological similarities and likely involve common mechanisms. Among the genes linked to PE, CHD2 has emerged as one of the most significant genetic predispositions. Galizia et al.7 observed that the overrepresentation of the unique CHD2 variant is associated with the typical photosensitive epilepsy syndrome, specifically absence seizures with eyelid myoclonia. A unique CHD2 variant was also found in photosensitivity generalized epilepsy patients. Furthermore, mutations in the CHD2 gene are linked to photosensitivity in certain epileptic encephalopathies, such as in some cases of Dravet syndrome.8
Another gene linked to photosensitivity is BRD2. Lorenz et al.5 found that variations in the BRD2 gene, located on chromosome 6p21, were associated with PPR. This gene has been shown to be involved in the mechanisms that cause epilepsy and other minor brain impairments, including abnormal brain wave activity as seen in EEG recordings9, 10 and photosensitivity.5, 6 Research on people of Caucasian German descent showed a relationship between genetic variations in the PPR and BRD2 genes, particularly in cases of photosensitive seizures. These findings suggest that BRD2 is a major gene influencing photosensitivity.5 Conversely, the investigation on Turkish descent failed to discover BRD2 gene mutations. It did not corroborate the prior presence of genetic variation shown to link with idiopathic photosensitive epilepsy.11 This conflicting result may vary due to racial genetic inheritance. Genetic linkages with epilepsy in the Indonesian population are still understudied, and no specific case of PE has been reported yet. The primary goal of this study was to ascertain whether a genetic polymorphism within the BRD2 gene is associated with an increased susceptibility to photosensitive epilepsy among individuals of Indonesian ancestry.
2 METHODS
2.1 Participants and study design
This was an observational study with a case–control design conducted at the Neurology Clinic of Universitas Sebelas Maret (UNS) Hospital, Sukoharjo, Indonesia, and the Biomedical and Biomolecular Laboratory, Faculty of Medicine, UNS, Surakarta, Indonesia. The inclusion criteria were patients of Indonesian descent with Javanese race. The exclusion criteria were significant neuropsychiatric disorders and a history of epileptic seizure (for the non-epilepsy group). This study used total sampling from May 2023 to May 2024 in accordance with the inclusion and exclusion criteria. A diagnosis of epilepsy and photosensitive epilepsy was made through the evaluation of clinical and neurophysiological data in accordance with the classification system promulgated by the International League Against Epilepsy (ILAE).
2.2 Diagnosis confirmation
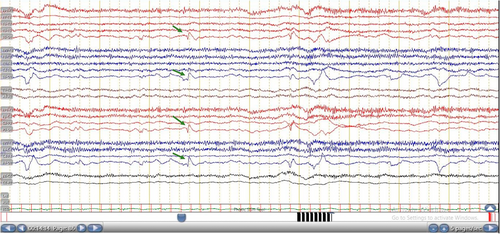
The patient was diagnosed with epilepsy, including photosensitive epilepsy, based on a thorough assessment encompassing medical history, physical examination, and electroencephalogram results. Reflex visual seizures were assessed during clinical evaluations and patient histories. Photosensitivity can be triggered by exposure to intermittent flashing lights, intense light, or sudden shifts between bright and dark conditions. Additionally, closing one's eyes or viewing patterns or television screens can induce photosensitive reactions due to the high-contrast stimuli they present.12 In this study, seizures were triggered by traffic lights and watching TV. Neurophysiological examination was performed by EEG using a standard intermittent photic stimulation (IPS) procedure.13, 14 Responses to IPS were categorized using the Waltz classification, and abnormal EEG responses were defined as PPR. Waltz and colleagues classified PPR into four distinct types, that is, type 1 (spikes first appear within the occipital lobe of the brain), type 2 (spikes occur in the area between parieto-occipital region, accompanied by slow wave with a specific biphasic pattern), type 3 (analogous to type 2, yet characterized by the propagation of neural spikes to the frontal cortical area), and type 4 (spikes and waves occur all over the brain or generalized).15-18 This study used frequencies within the range of 15–20 Hz, which are particularly relevant for inducing PPR during EEG testing.1 Two epileptologists have conducted single-blind investigations on EEG data. In cases where there was discordance between the two epileptologists, a consensus meeting was held to resolve discrepancies. We found discordance in classifying PPR types from one case, which was successfully addressed through the consensus process.
2.3 Polymorphism confirmation
The BRD2 gene (BRD2 bromodomain containing 2 (Homo sapiens [human]); location: 6p21.32 – NC_000006.12 (32968594…32981505); exon count: 17; assembly: GRCh38.p14 (GCF_000001405.40)) by NCBI was chosen in this study. BRD2 is located on chromosome 6 (Homo sapiens chromosome 6, GRCh38.p14 Primary Assembly; NCBI reference sequence: NC_000006.12; length: 170805979 bp; assembly: GCF_000001405.40). The genotypes and alleles of three single nucleotide polymorphisms (SNP1-3) covering the BRD2 sequence were investigated for each study participant. The rs206781 and rs188245 BRD2 polymorphisms were found to have strong allelic and haplotypic associations with PPR in German populations5 and rs15912 is a BRD2 gene variation with the highest number of samples in Turkish populations.11
Blood specimens were obtained via venipuncture of the median cubital vein and placed into EDTA-treated vacutainer tubes (BD Biosciences, USA). DNA was isolated from peripheral blood mononuclear cells (PBMCs), yielding an approximate concentration of 50 ng/L. Subsequent investigative procedures incorporated the utilization of polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) for the analysis of DNA. Laboratory technicians isolated DNA from a sample of the subject's white blood cells using the Promega® isolation kit. Next, the DNA was amplified using the Master Mix Go Taq Green Promega® kit. The primer to detect the rs206781, rs188245,5 and rs1591211 SNPs was developed using SnapGene software (Dotmatics, USA) and is displayed in Table 1. BRD2 promoter amplification (rs206781, rs188245, and rs15912) used the PCR method with its primers. The PCR conditions used included pre-denaturation, amplification, denaturation, annealing, and extension following the laboratory user manual. The PCR product was then cut with the BfaI enzyme (MBI Fermentas®). The DNA amplification results were electrophoresed on a 3% agarose gel in Tris-Boric Acid-EDTA buffer (Tris Borate 0.045 M and ethylene diamine tetra acetic acid 0.001 M pH 8.0) containing ethidium bromide and using a Horizontal Mini Sub DNA electrophoresis apparatus (Bio-Rad Lab). Subsequently, the results of BRD2 gene amplification were cloned in the private company PT. Genetika Science, Indonesia. They underwent a sequencing process using the Sanger sequencing method at the 1st Base Laboratory, Apical Scientific, Selangor, Malaysia. The obtained sequences were checked for polymorphisms using the SnapGene software (Dotmatics, USA).
| Polymorphism number | dbSNP | Alleles | Position | Primer | Product size (bp) |
|---|---|---|---|---|---|
| SNP1 | rs206781 | C/T | chr6:32978356 (GRCh38.p14) |
|
437 bp |
| SNP2 | rs188245 | A/G | chr6:32988199 (GRCh38.p14) |
|
166 bp |
| SNP3 | rs15912 | C/G | chr6:32976317 (GRCh38.p14) |
|
135 bp |
- Abbreviations: bp, base pair; rs, restriction site; SNP, single nucleotide polymorphism.
2.4 Statistical analysis
Statistical analyses were carried out on a Mac platform utilizing SPSS software version 27.0 (IBM Corp.). Demographic characteristics, genotype, and allelic frequencies of the SNP were compared between these two groups using the statistical method of Chi-square (χ2) tests. The findings were statistically significant, with a p < 0.05.
3 RESULTS
Data were collected from 27 individuals for this study. As participant characteristics (demographic and clinical data) shown in Table 2, the data did not demonstrate statistically significant differences among the three groups of participants with respect to sex, age, seizure type, disease duration, or medication. All epilepsy patients received valproic acid (VPA) 500–750 mg daily with a median dosage of 600 mg a day.
| Characteristics | Epilepsy group | Non-epilepsy group, n (%), (N = 10) | p-Value | |
|---|---|---|---|---|
| PE (+), n (%), (N = 9) | PE (−), n (%), (N = 8) | |||
| Sex | ||||
| Male | 4 (44.4%) | 3 (37.5%) | 4 (40.0%) | 0.957 |
| Female | 5 (55.6%) | 5 (62.5%) | 6 (60.0%) | |
| Age | ||||
| <18 years | 4 (44.4%) | 4 (50.0%) | 4 (40.0%) | 0.982 |
| 18–60 years | 3 (33.3%) | 3 (37.5%) | 4 (40.0%) | |
| >60 years | 2 (22.2%) | 1 (12.5%) | 2 (20.0%) | |
| Seizure types | ||||
| Generalized tonic–clonic | 5 (55.6%) | 4 (50.0%) | 0.819 | |
| Focal | 4 (44.4%) | 4 (50.0%) | ||
| PPR Type (15–20 Hz) | ||||
| Type 1 | 6 (66.7%) | 0.503 | ||
| Type 2 | 1 (11.1%) | |||
| Type 3 | 1 (11.1%) | |||
| Type 4 | 1 (11.1%) | |||
| Disease duration | ||||
| <5 years | 2 (22.2%) | 3 (37.5%) | 0.745 | |
| 5–10 years | 5 (55.6%) | 4 (50.0%) | ||
| >10 years | 2 (22.2%) | 1 (12.5%) | ||
| History of medication | ||||
| Valproic acid | 9 (100.0%) | 8 (100.0%) | ||
- Abbreviation: PE, photosensitive epilepsy.
In the PE group, 66.7% of patients exhibited a Waltz type 1 response to IPS, suggesting an occipital spike response (Figure 1) rather than a robust epileptogenic PPR (Waltz types 2–4). However, these patients also exhibited interictal epileptiform discharges in other epochs, which raised suspicion of an abnormality potentially linked to epileptiform discharges.
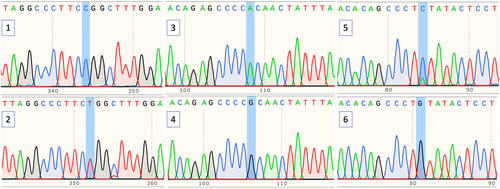
BRD2 (rs206781, rs188245, and rs15912) SNPs sequencing in 27 patients was conducted (Figure 2). This study revealed a significant statistical difference between genotype (rs206781, p = 0.008 and rs188245, p = 0.004) and allele frequencies (rs206781, p = 0.000 and rs188245, p = 0.000) for the BRD2 gene between groups (Table 3).
| Polymorphism | Epilepsy group | Non-epilepsy group, n (%), (N = 10) | p-Value | |
|---|---|---|---|---|
| PE (+), n (%), (N = 9) | PE (−), n (%), (N = 8) | |||
| SNP 1 (rs206781) | ||||
| Genotypes | ||||
| CC | 0 (0.0%) | 5 (62.5%) | 7 (70.0%) | 0.008* |
| CT | 4 (44.4%) | 2 (25.0%) | 3 (30.0%) | |
| TT | 5 (55.6%) | 1 (12.5%) | 0 (0.0%) | |
| Alleles | ||||
| C | 4 (22.2%) | 12 (75.0%) | 17 (85.0%) | 0.000* |
| T | 14 (77.8%) | 4 (25.0%) | 3 (15.0%) | |
| SNP 2 (rs188245) | ||||
| Genotypes | ||||
| AA | 0 (0.0%) | 6 (75.0%) | 6 (60.0%) | 0.004* |
| AG | 3 (33.3%) | 2 (25.0%) | 3 (30.0%) | |
| GG | 6 (66.6%) | 0 (0.0%) | 1 (10.0%) | |
| Alleles | ||||
| A | 3 (16.7%) | 14 (87.5%) | 15 (75.0%) | 0.000* |
| G | 15 (83.3%) | 2 (12.5%) | 5 (25.0%) | |
| SNP 3 (rs15912) | ||||
| Genotypes | ||||
| CC | 3 (33.3%) | 2 (25.0%) | 3 (30.0%) | 0.506 |
| CG | 6 (66.6%) | 4 (50.0%) | 4 (40.0%) | |
| GG | 0 (0.0%) | 2 (25.0%) | 3 (30.0%) | |
| Alleles | ||||
| C | 12 (66.7%) | 8 (50.0%) | 10 (50.0%) | 0.509 |
| G | 6 (33.3%) | 8 (50.0%) | 10 (50.0%) | |
- Abbreviation: PE, photosensitive epilepsy.
- * p < 0.05 (considered statistically significant).
4 DISCUSSION
Although substantial strides have been achieved, the intricate biological processes that contribute to photosensitive epilepsy continue to perplex researchers. However, aberrant synchronization of neural activity is believed to be involved. Although it is known that photosensitive epilepsy is a cerebral phenomenon, the inheritance is intricate and involves multiple genes that likely interact. The investigations of molecular genetics have thus far identified probable loci on chromosomes 2, 6, 7, and 16.6, 19, 20 Previous research has identified genetic relationships between specific chromosomal regions and different types of epilepsy. The 7q32 and 16p13 regions have been linked to PPR families predominantly affected by myoclonic seizures,19 whereas the 6p21 and 13q31 loci have been identified as potential genetic risk factors for absence and partial seizure phenotypes within PPR kindreds.6 This study confirmed the association of BRD2 polymorphism (rs206781 and rs188245) with PE. This study aligns with Lorenz et al., who also found a relationship between these SNPs. Lorenz et al.5 identified a correlation between specific genetic markers within the BRD2 region and the development of PPR in a German population. Interestingly, a considerable number of these patients (59%) were additionally diagnosed with one or more idiopathic generalized epilepsy (IGE) syndromes, including Juvenile Absence Epilepsy (JAE), Childhood Absence Epilepsy (CAE), Juvenile Myoclonic Epilepsy (JME), Epilepsy with Generalized Motor Aggravation (EGMA), and Epilepsy with Generalized Tonic–Clonic Seizures (EGT). Meanwhile, other studies yielded different results; Yavuz's research did not confirm the existence of specific genetic variations in the BRD2 gene, which were previously recognized as factors contributing to idiopathic photosensitive epilepsy. On the other hand, his study showed a clear and statistically significant relationship between specific genetic variations within the BRD2 gene (known as single nucleotide polymorphisms) and the way photosensitive epilepsy develops and evolves in affected individuals.11 The rs206781 and rs188245 BRD2 polymorphisms were found to have strong allelic and haplotypic associations with PPR in German populations,5 and rs15912 is a BRD2 gene variation with the highest number of samples in Turkey populations.
Genetic studies have shown a connection between the 6p21 region of chromosome 6 and IGE, with a particular focus on its role in JME and PE.10, 21 The involvement of the BRD2 gene in JME/PPR has been suggested by several lines of investigation, which exhibit a range of genetic differences. The BRD2 gene, a potential regulator of gene expression, is a strong candidate for the genetic cause of JME. Research by Pal et al.22 has identified specific genetic variations in the BRD2 gene, that is, two SNP variations in the RING3 promoter area that were associated with JME. Furthermore, studies have linked changes in the BRD2 gene to alterations in the brain's GABA system, which regulates seizures in IGE.23 The BRD2 gene exhibits tissue-specific expression patterns, generating transcripts with diverse 50 untranslated regions (UTR) lengths. The presence of an alternatively spliced, highly conserved exon can prematurely terminate translation. The intricate regulatory landscape governing BRD2 protein synthesis within the developing brain may contribute to an increased likelihood of seizure activity.24
It remains unknown what role BRD2 has in epileptogenesis. The brain synthesizes BRD2, a nuclear transcriptional regulator that is part of the family of proteins called BET. Characteristically, the BRD2 protein exhibits two bromodomains. These domains fulfill a critical role in the recognition and binding of acetylated lysine residues situated within histone tails. The capacity to identify and firmly attach to locations within the genome directly influences the dynamic changes in chromatin organization. This process of chromatin remodeling is fundamental, as it regulates the activation or suppression of genes and other vital cellular processes.25, 26 Employing a modified fluorescence resonance energy transfer approach on a flow cytometer, in the context of the living cell nucleus, BRD2 has been shown to selectively engage with histone H4 molecules that are posttranslationally modified by acetylation at lysine 12.25 The function of BRD2 in promoting gene transcription was contingent upon its interaction with histone proteins bearing acetyl modifications. Furthermore, its connection to acetylated chromatin was maintained on chromosomes even as the cell underwent mitosis.25 BRD2 might regulate gene expression patterns and potentially establish a biological process that allows epigenetic transcriptional memory to be inherited by daughter cells.25, 26 As a result, BRD2 may play a part in the developmental processes that shape the nervous system and determine cell types. Moreover, this might contribute to the hypoplasia of mesiofrontal cortical structures, which is a frequent neuroanatomical observation in patients with JME and other IGE.27, 28
Even though these studies provide significant insights into the genetic foundation of PE, it is crucial to acknowledge that the connection between specific genes and this disorder is intricate and multifaceted. Additional research is necessary to achieve a thorough understanding of the underlying mechanisms. Multiple genes have been related to photosensitive epilepsy in diverse genetic studies. Genetic studies have implicated CHD2 and SCN2A in the pathogenesis of PE. CHD2 encodes a chromatin-remodeling protein that regulates gene expression. Several neurological diseases, including epilepsy, have been related to mutations in CHD2.29 SCN2A encodes a voltage-gated sodium channel required for initiating and propagating action potentials in neurons. Several epilepsy disorders, including photosensitive epilepsy, have been related to mutations in SCN2A.30 GABRA1 encodes a subunit of the gamma-aminobutyric acid type A (GABA-A) receptor, a crucial neurotransmitter in regulating seizures.31, 32 SLC2A1 encodes a glucose transporter that is essential for brain energy supply. Several epilepsy disorders, including photosensitive epilepsy, are related to SLC2A1 mutations.33 In addition, the 6p21 locus encompasses a significant array of genes that could potentially contribute to an increased risk of epilepsy, for example, GABBR1, GRM4, KCNK16, KCNK17, and ALDH5A1.6, 34-37
All epilepsy patients enrolled in this study were administered VPA as an anti-seizure medication. Patients with focal seizure symptoms were also already taking VPA when they were referred to our hospital from primary health care facilities. The use of VPA was due to some clinical reasons, that is, phenytoin-induced gingival hyperplasia or drug-induced allergic reaction due to carbamazepine.
The dosage of VPA received by our patients sometimes did not meet the minimum dosage suggested for photosensitive epilepsy, that is, 2000 mg a day,16 although the patients had controlled seizures, the reaction to IPS might not have been suppressed by the VPA use, as shown by the EEG recordings. This dosage was restricted due to the national universal health coverage policy since all the patients were covered by national health insurance from the Indonesian government.
While the incidence of photosensitivity is well documented in Western populations, with studies estimating a prevalence of one case per 100 000 of all new epilepsy cases globally,38 there is no data on its prevalence in Indonesia. In our center, from 687 EEGs performed on patients with epilepsy during the same period as this study, only nine exhibited a PPR (1.3%), which emphasizes that photosensitivity, though affecting a minority of our population, presents a clinically relevant concern. The present findings point toward the potential of BRD2 polymorphisms to function as a genetic marker for PE in the Indonesian population. Nonetheless, the limited prevalence of PPR within our EEG dataset suggests that additional genetic and environmental variables may impact the manifestation of photosensitivity.
4.1 Limitations
It should be mentioned that this study has several limitations. This study was performed in only one race in Indonesia, which is Javanese. Subsequently, conventional PCR was used to detect SNPs, which is not the most advanced technique. Therefore, it is suggested that additional research be conducted using a multiracial approach and utilize the most recent direct sequencing approach with a cohort study design. Evaluating other SNP locations or genes, such as rs206786, rs3918149, rs2071571, rs516535, rs206777,5 rs9276935, rs55912052, rs516535, rs15912, rs2071876, rs3918140,11 as well as other genes (for instance CHD2, SCN2A, GABRA1, and SLC2A1) is also suggested. It is anticipated that investigations will discover possibly key elements and the etiologies of the development of photosensitive epilepsy. Additional imaging and functional investigations, such as fMRI and MEG studies, may also be undertaken to elucidate pathophysiology.
5 CLINICAL RELEVANCE OR FUTURE DIRECTIONS
Our study confirms that the genetic variations of BRD2 (rs206781 and rs188245) are associated with PE in Indonesian descendants of the Javanese race. The findings of this study corroborate prior research indicating a potential correlation between BRD2 gene polymorphisms and photosensitive epilepsy. To acquire a complete knowledge of the development of photosensitive epilepsy, further polymorphism studies at other SNP locations or genes are necessary.
6 STATISTICAL METHODS
6.1 Analysis guidelines
The data were analyzed with the assistance of SPSS Statistics version 27.0 for Mac, a software package developed by IBM Corporation in the United States. Demographic characteristics, genotype, and allelic frequencies of the SNP were compared between both groups using the Chi-square (χ2) tests. Exact methods were utilized for the analysis of categorical data to ensure precision. A threshold of p < 0.05 was used to assess statistical significance.
6.2 Presentation guidelines
Two-sided p-values were presented. For p-values exceeding 0.01, precision was generally reported to two decimal places, with an exception for values between 0.01 and 0.001, which were displayed to three decimal places. p-values below 0.001 were indicated as less than 0.001.
AUTHOR CONTRIBUTIONS
Diah Kurnia Mirawati: Study conceptualization and design, and data collection. Muhana Fawwazy Ilyas: Data collection, statistical analyses, and manuscript preparation. Muhammad Hafizhan: Study conceptualization and design, data collection, and manuscript preparation. Stefanus Erdana Putra: Study conceptualization and design, data collection, and manuscript preparation. Hanindia Riani Prabaningtyas: Study conceptualization and design, and data collection. Pepi Budianto: Study conceptualization and design, and data collection. Suroto Suroto, Subandi Subandi, Rivan Danuaji, Yetty Hambarsari, and Baarid Luqman Hamidi: Data validation and supervision. Ervina Arta Jayanti Hutabarat, Ira Ristinawati, Teddy Tejomukti, Raden Andi Ario Tedjo, and Faris Khairuddin Syah: Data validation and visualization.
ACKNOWLEDGMENTS
No medical writer or editor was involved in the creation of our manuscript.
CONFLICT OF INTEREST STATEMENT
None of the authors has any conflict of interest to disclose.
ETHICAL APPROVAL
The Health Research Ethics Committee of Universitas Sebelas Maret granted approval to the study protocol with the number 588/IV/HREC/2023, which was approved on April 16, 2023. All participants signed written informed consent. We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Open Research
DATA AVAILABILITY STATEMENT
Access to the data that corroborates the research results is available from the corresponding author upon inquiry.