Gut commensal microbes do not represent a dominant antigenic source for continuous CD4+ T-cell activation during HIV-1 infection
Abstract
Chronic immune activation is a hallmark of HIV-1 infection; specifically, the activation of T cells has predictive value for progression to AIDS. The majority of hyperactivated T cells are not HIV-specific and their antigenic specificities remain poorly understood. Translocation of gut luminal microbial products to systemic sites contributes to chronic immune activation during HIV-1 infection, but how it affects (TCR-dependent) immune activation remains elusive. We hypothesized that gut luminal antigens foster activation of CD4+ T cells with specificities for commensal bacterial antigens, thereby contributing to the pool of activated CD4+ T cells in the circulation of HIV-1 infected individuals. To test this hypothesis, we quantified the frequencies of gut microbe-specific CD4+ T cells by cytokine production upon restimulation with selected gut commensal microbial antigens. Contrary to our hypothesis, we did not observe increased but rather decreased frequencies of gut microbe-specific CD4+ T cells in HIV-1 infected individuals compared to healthy controls. We conclude that the increased activation status of circulating CD4+ T cells in HIV-1 infected individuals is not driven by CD4+ T cells with specificities for commensal bacterial antigens.
Introduction
Chronic activation of the immune system is a major determinant of progression to AIDS 1 and particularly the level of hyperactivated T cells is considered as best prognostic marker for disease progression 2. HIV-1 infection and replication is the main driving force of persistent, pathological level of T-cell activation 1, 3 but HIV-1 specific T cells account for only a small fraction of activated T cells 4-6, implicating substantial “bystander” activation. Multiple mechanisms are considered to contribute to the genesis of such extensive immune activation, many of them are related to HIV-1 associated immunodeficiency 7, 8, sustained inflammation 1, and microbial translocation 9.
HIV-1-related immunodeficiency leads to the reactivation of ubiquitous symbiotic viruses (e.g. herpes viruses), generating HIV-unrelated antigens 7, 8. Furthermore, the rapid and irreversible depletion of intestinal CD4+ T cells, in particular Th17 cells that are essential for maintaining barrier function 10, 11 during HIV-1 infection impair the integrity of the intestinal barrier and enable the translocation of commensal microbes (or their products) from the gut lumen to systemic sites 9. Thus, HIV-1 replication, HIV-1-associated immunodeficiency 7, 8, and microbial translocation 9 fuel a milieu rich of viral and bacterial pathogen-associated molecular patterns and proteins that have the capacity to activate innate and adaptive immune cells by engaging pattern recognition receptors and rearranged high affinity antigen receptors, respectively. Indeed, constant triggering of these receptors in combination with the ensuing cytokine secretion likely all contribute to HIV-1 associated hyper-activation of the immune system, which is best described for T cells. Interestingly, causes for activation of non-HIV-specific CD4+ and CD8+ T cells seem to differ with respect to involvement of TCR engagement: whereas CD8+ T cells can easily be activated by cytokines (e.g. IL-15) in a TCR-independent manner 12, the activation of CD4+ T cells seems to depend more on TCR-dependent cognate antigen sensing 13, 14. DCs, among other cells, adopt a more mature and activated phenotype 13, 15, 16, promoting TCR-mediated CD4+ T-cell activation, in particular in situations of low antigen abundance. Nevertheless, the antigenic specificities of non HIV-specific hyperactivated CD4+ T cells remain poorly defined. We have previously demonstrated that HIV-1 replication leads to the activation of herpes virus-specific CD4+ T cells even in the absence of detectable herpes virus replication 13, 17. However, herpes virus-specificity only accounts for a small fraction of non-HIV-specific activated CD4+ T cells during HIV-1 infection. Here, we hypothesized that activated CD4+ T cells might also compromise specificities for gut microbial antigens, based on the increased exposure of systemic immune cells to gut commensal microbes 9. To test this hypothesis, we measured the frequencies of circulating gut-microbe specific CD4+ T cells with specificity for selected gut commensal microbial antigens. We included not only HIV-1 patients but also inflammatory bowel disease (IBD) patients to compare to another condition of sustained gastrointestinal inflammation 18, 19.
Results and discussion
CD4+ T cells specific for gut commensal microbes are present in blood of healthy individuals
The structural and immunological integrity of the intestinal barrier is crucial for the confinement of commensal microbes to the gut lumen and for a strict compartmentalization between the gastrointestinal and systemic immune system—at least in clean experimental conditions such as specific-pathogen free mice 20. In contrast, humans are repeatedly experiencing episodes of compromised gastrointestinal barrier integrity, provoked by, e.g. gastrointestinal tract (GIT) infections and behavioral patterns affecting the integrity of the GIT barrier function 21. These episodes are associated with the exposure of gut microbial products to immune cells at systemic sites, documented by serum IgG antibodies with specificities for gut microbial antigens being present in healthy donors 22, 23. It is unclear whether this also holds true for CD4+ T cells with gut microbial specificities. Hence we investigated this question by analyzing cytokine production upon restimulation with lysates from gut commensal microbes.
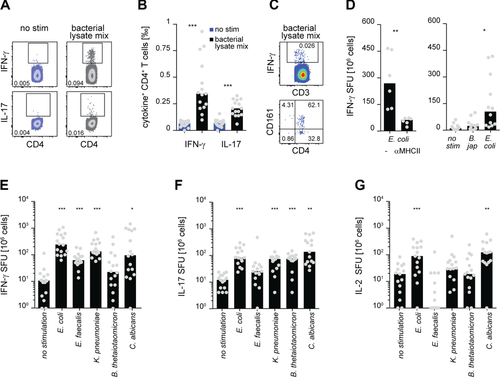
Indeed, CD4+ T cells isolated from blood produced IFN-γ and IL-17 upon restimulation with a mixture of Escherichia coli (E. coli), Klebsiella pneumoniae (K. pneumoniae), Enterococcus faecalis (E. faecalis), and Bacteroides thetaiotaomicron (B. thetaiotaomicron) derived bacterial lysates in healthy individuals, albeit at very low frequencies (Fig. 1A and B). As only very few of the bacterial lysate-responsive IFN-γ producing cells were CD4 negative and expressed CD161 (identifying invariant Mucosal-associated Invariant T cells), we conclude that CD4+ T cells are the predominant source of these cytokines (Fig. 1C). Furthermore, the responses were dependent on TCR-MHC interaction. MHC II blocking during restimulation with E. coli lysate led to significant reduction of IFN-γ production (Fig. 1D, left). Stimulating CD4+ T cells with bacterial lysates could include responses specific for conserved intracellular bacterial proteins that are shared between many bacterial species and therefore not being an identifier for gut commensals. Since CD4+ T cells produced much less IFN-γ upon restimulation with a lysate derived from the non-human-related soil bacterium B. japonicum compared to E. coli lysate, we conclude that gut microbe-specific CD4+ T cells are indeed found in the circulation of healthy humans and that our experimental approach is able to detect those responses (Fig. 1D, right). Restimulation of CD4+ T cells with lysates derived from E. coli, K. pneumoniae, and C. albicans revealed relatively high frequencies of IFN-γ, IL-17, and IL-2 producing CD4+ T cells (Fig. 1E–G), consistent with previous reports documenting systemic IgG responses against gut commensal microbes in the serum of healthy individuals 22, 23. Together, these data imply that the strict compartmentalization between gut mucosal and systemic immune responses toward commensal microbes is—at least to some extent—broken in healthy human individuals most likely in response to transient episodes of bacterial translocation 21. Because CD4+ T-cell responses specific for E. faecalis and B. thetaiotaomicron were generally close to the detection limit, we did not include these lysates in the further experiments.
Gut microbe-specific CD4+ T cells do not contribute to activated T cells during HIV-1 infection
The intestinal barrier integrity is severely impaired in HIV-1 infected individuals, allowing translocation of gut commensal luminal products into the systemic circulation 9. We therefore examined whether this is associated with higher frequencies of systemic CD4+ T cells with gut microbial specificities in HIV-1 patients (Supporting Information Table 1). To our surprise, the frequencies of INF-γ producing CD4+ T cells specific for E. coli, K. pneumoniae, and C. albicans were significantly lower in HIV-1 patients compared to healthy controls (Fig. 2A) and a similar trend toward lower frequencies of E. coli-, K. pneumoniae-, and C. albicans-specific CD4+ T cells producing IL-17 and IL-2 was observed (Fig. 2B and C).
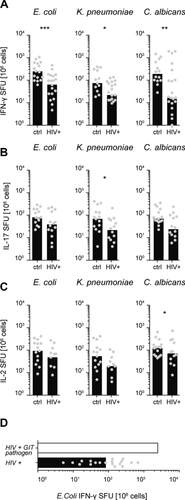
These results indicate that gut microbe-specific CD4+ T cells—despite presumably prolonged availability of gut microbial antigens at systemic sites during HIV-1 infection—are not a major constituent of activated CD4+ T cells in the blood of HIV-1 patients. This is likely caused by several factors: during active HIV-1 replication gut commensal-specific CD4+ T cells might be targets for infection which might curtail their abundance. We therefore deliberately enrolled HIV-1 infected patients on successful antiretroviral combination therapy (cART) for this analysis (Supporting Information Table 1), reflected by relative low levels of systemic immune activation as determined by coexpression of HLA-DR and CD38 on peripheral blood T cells (Supporting Information Fig. 1) and CD4 T-cell counts were restored to relative high levels (Supporting Information Table 1). Furthermore, CD4+ T-cell responses specific for E. coli (Fig. 2D) were highly elevated in a patient exhibiting a GIT infection at the time of blood withdrawal, likely reflecting a transient decrease of the intestinal barrier function along with expansion of microbiota-specific T cells 24. This observation suggests that immune competence in HIV-1 infected individuals is not compromised to an extent that it would prevent priming of gut microbe-specific CD4+ T-cell responses. Another potential factor that might substantially curtail the frequencies of gut-microbe-specific CD4+ T cells at systemic sites in HIV-1 patients is the severe and pronounced depletion of intestinal CD4+ T cells 10, 11 whose recovery is not achieved with cART 25.
IBD patients exhibit comparable levels of gut commensal-specific CD4+ T cells as healthy controls
IBD patients suffer from sustained gastrointestinal tract inflammation that compromises intestinal barrier function and is associated with pronounced microbial translocation 18, 19. Importantly, this pathology is accompanied by local but not systemic immune activation and thereby represents an ideal condition to analyze whether the presence of gut luminal products in the circulation translates into high levels of systemic CD4+ T cells with corresponding specificities. Along that line, we and others have demonstrated that the augmented exposure of systemic immune cells to constituents of the gut commensal microbiota leads to enhanced levels of systemic microbe-specific antibody responses 22, 23. Interestingly, peripheral CD4+ T cells from IBD patients were not substantially enriched for specificities toward gut commensal microbes (Fig. 3A–C), except the frequency of IL-2 producing CD4+ T cells specific for E. coli were slightly elevated in IBD patients compared to healthy controls.
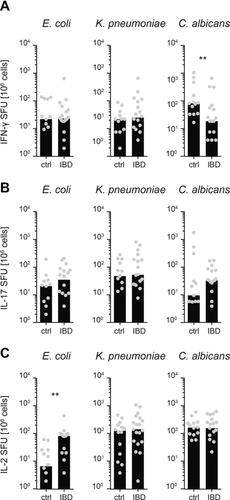
Excessive GIT homing of CD4+ T cells is observed in IBD-associated pathology 26 and could explain why no increased frequencies of gut microbe-specific CD4+ T cells are systemically detectable in IBD patients compared to healthy controls. Such excessive gut homing does probably not explain the absence of gut microbe-specific CD4+ T cells in the circulation of HIV-1 patients, since gut homing of CD4+ T cells is reported to be impaired in HIV-1 patients 27. Furthermore, our own findings suggest that bacteria-specific IL-17 producing CD4+ T cells do not express high levels of gut homing receptors α4β7 (at least in healthy donors) (Supporting Information Fig. 2), what is in contrast to C. albicans- or CMV-specific cells and hence rather speaks against gut homing as major contributor for depleting the pool of bacteria-specific CD4+ T cells from the circulation. Unfortunately, we did not have the chance to investigate gut biopsies from these patients to corroborate this statement. Since IBD patients were treated with anti-TNF-α (Infliximab) that was reported to downregulate INF-γ production by T cells 28, 29, we cannot exclude that we underestimated the frequencies of gut microbe-specific CD4+ T cells based on their ability to produce IFN-γ upon in vitro restimulation. Also, repetitive stimulation of gut-microbe-specific CD4+ T cells by bacterial antigens in the context of microbial translocation 30 could induce their functional impairment that might explain why those cells cannot be detected upon restimulation with the corresponding bacterial lysates.
Concluding remarks
In summary, our study shows that there are low frequencies of gut microbe-specific CD4+ T cells in the periphery of HIV-1 patients and there are likely a number of factors precluding accumulation of those cells in circulation despite microbial translocation and enteropathy. However, we can conclude that gut commensal microbe specificities are not effectively contributing to the peripheral population of activated CD4+ T cells and thereby our study excludes gut commensal derived cognate antigens as major driving force for TCR-dependent CD4+ T-cell activation.
Materials and methods
Patients
HIV patients (N = 22) were enrolled in the Zurich Primary HIV Infection Study (www.clinicaltrials.gov; ID: NCT00537966) 31 who initiated cART treatment within 90 days after acute infection. Patients chosen for our study experienced an interruption of therapy for at least 1 year, cART had been resumed on average 6 years before blood samples were drawn for the present study and no viral replication was detectable at that time point 32, 33. Patients with clinically and histologically documented IBD (N = 19) were enrolled by the Swiss IBD cohort study (http://ibdcohort.ch), only patients receiving Infliximab (anti-TNF-α) were included in this study. Patients on immunosuppressive therapies were excluded.
Healthy individuals of comparable age were recruited at the same time. Approval and written informed consent from all patients were obtained according to the guidelines of the Ethics Committee of the University Hospital Zurich and the Triemli Hospital.
Bacterial and Candida lysate
Overnight cultures of E. coli, K. pneumoniae, and E. faecalis (primary isolates from stool samples of healthy individuals), B. thetaiotaomicron (gift from Dr. Emma Slack), B. japonicum (gift from Prof. Dr. Hauke Hennecke), and C. albicans strain SC5314 34 (gift from Prof. Dr. Salomé Leibundgut-Landmann) were adjusted to 1 × 108 cells/mL in PBS. Bacteria were sonicated and incubated on ice alternately for 1 min, C. albicans was boiled for at least 1 h. Lysate concentration used for the stimulation of CD4+ T cells corresponded to 1 × 105 cells/mL. Lysates were stored frozen at –20°C or kept at 4°C if used within a couple of weeks.
Enumeration of cytokine producing CD4+ T cells
CD4+ T cells and CD14+ monocytes were isolated from PBMCs using human CD4 and -CD14 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instruction. CD4+ T cells were stimulated with bacterial and candida lysates or 15-mer peptides covering the entire immunodominant CMV pp65 protein or IE-1 in the presence of freshly isolated autologous CD14+ monocytes in a ratio of 4 to 1 for 18 h at 37°C. A total of 2 – 2.5 × 105 CD4+ cells were used in enzyme-linked immuno spot (ELISpot) assays, whereas 1 × 106 CD4+ cells were used for analysis by FACS. MHCII-specific Ab (clone TÜ39; f.c. 2.5 μg/mL, BioLegend, Switzerland) or brefeldin A (f.c. 10 μg/mL, Sigma, Switzerland) was added if required. For assay controls, CD4+ T cells were left unstimulated or stimulated with PMA & ionomycin (50 and 500 ng/mL, Sigma, Switzerland). The assay was performed in duplicates in a total volume of 200 μL of RPMI-1640 supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine (all reagents from PAA Laboratories, Austria). Detection of antigen-specific CD4+ T-cell responses by ELISpot (Mabtech, Sweden) was performed according to the manufacturer's instructions using MAIPSWU plates (Millipore, Darmstadt, Germany) and spots were counted on an AID ELISpot reader (CTL, Germany). For the analysis of intracellular IL-17 and IFN-γ expression, cells were permeabilized after surface staining with Permeabilization buffer (BioLegend, San Diego, USA) and stained for 30 min at RT with fluorchrome conjugated monoclonal antibodies (anti-IL-17: clone BL168; anti-IFN-γ: clone B27). Surface staining was performed with fluorchrome conjugated monoclonal anti-CD3 (clone UHCT-1), anti-CD4 (clone SK3), anti-CD38 (clone HIT-2a), anti-HLA-DR (clone L243), anti-integrin β antibody (clone FIB504), and CD161 (clone HP-3G10) for 30 min at 4°C. LIVE/DEAD cell stain (Life Technologies, Carlsbad, USA) was used for the exclusion of dead cells. LSR II and FACSDiva software (BD Biosciences, Franklin Lakes, USA) were used for the acquisition of flow cytometry data, and FlowJo software (TreeStar) was used for analysis.
All antibodies were purchased from BD Biosciences, Switzerland, or BioLegend, San Diego, USA). Data were analyzed and plotted with GraphPad Prism (GraphPad, La Jolla, CA, USA) using scatter plots with bars representing median values.
Acknowledgments
We are grateful to the patients who agreed to donate blood to support our experiments and we especially thank the study nurses and doctors in the Triemli hospital as well as at the department of infectious disease at the University hospital in Zurich for their extra efforts. A special thank goes to Dr. med Dominique Braun and Christina Grube for excellent patient care within the ZPHI study. We are grateful to Dr. Peter Mirtschink who took blood from healthy donors. We thank Dr. Hauke Hennecke for providing us with B japonicum and Dr. Salomé LeibundGut-Landmann for providing us with C. albicans. This work was supported by the ETH Zurich, the Swiss National Science Foundation (Grant No. 310030_129751 to A.O., by the University of Zurich's Clinical research Priority Program (CRPP) “Viral infectious diseases: Zurich Primary HIV Infection Study” to H.F.G.) and the Horten Foundation. Thanks to the Institute of Medical Microbiology, University of Zurich, for the isolation and typefication of gut commensal bacteria derived from primary stool samples.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
References
Abbreviations
-
- cART
-
- antiretroviral combination therapy
-
- ELISpot
-
- enzyme-linked immuno spot assay
-
- GIT
-
- gastrointestinal tract
-
- IBD
-
- Inflammatory bowel disease