CRISPRi screening identifies PIKfyve as a co-therapeutic target for obinutuzumab
Dear Editor:
Combining monoclonal antibodies with targeted agents is a promising yet mechanistically underexplored strategy for B-cell lymphoma therapy.1 Through a CRISPR interference (CRISPRi) screen, we identify PIKfyve as a suppressor of obinutuzumab-induced lysosomal membrane permeabilisation (LMP) and direct cell death (DCD), highlighting its potential as a combinatorial therapeutic target.
Obinutuzumab (OBI), a next-generation anti-CD20 monoclonal antibody, is known to trigger DCD via LMP in B-cell malignancies.2 However, the cellular factors that regulate this pathway remain poorly defined.3 To systematically identify modulators of OBI-induced cytotoxicity, we conducted a customised CRISPRi screen in Raji B cells targeting 2318 druggable genes4 (Figure 1A). Following iterative rounds of OBI treatment and selection, next-generation sequencing (NGS) analysis revealed genes whose knockdown either sensitised cells to or conferred resistance against OBI-induced LMP and DCD (Figure 1B). Using Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK, Table S1), we ranked genes based on their depletion or enrichment following OBI treatment, identifying key regulators of LMP and DCD (Figure 1C). Essential genes critical for cell survival were excluded, as their sgRNAs were predominantly depleted. To prioritise clinically relevant targets, we applied a selection strategy: (1) applying FDR cutoff of 25%, (2) excluding essential genes based on the DepMap database and Raji B-cell essential gene list, specifically selecting genes with DepMap gene essentiality scores < –1 and at least three high-quality sgRNAs (resulting in 97 genes, Table S2), and (3) focus on druggable targets with available inhibitors. This yielded 15 candidate genes for further validation. The sgRNA enrichment profiles and their relative importance are summarised in a box plot (Figure 1D and E).

To validate this finding, we generated single-knockdown Raji cell lines for 14 candidate genes and confirmed their knockdown efficiency (Figure 2A, Tables S3 and S4; CDK12 was excluded due to the inability to establish a stable knockdown line). Among these, six genes (DCTD, SMS, ATP1A1, SLC16A1, ODC1, and PIKfyve) significantly increased LMP and DCD, reinforcing their potential as modulators of lysosomal disruption (Figure 2B–D). Apilimod, a selective PIKfyve inhibitor, showed the strongest enhancement of OBI-induced DCD with minimal cytotoxicity alone (Figure 2E and F). Additionally, Apilimod dose-dependently enhanced DCD induced by OBI and RTX, indicating that PIKfyve inhibition broadly amplifies anti-CD20-mediated cytotoxicity (Figure 2G–J). Further, Umbralisib (a PI3Kδ inhibitor) enhanced OBI-induced cytotoxicity, whereas Acalabrutinib (a BTK inhibitor) had no effect, suggesting that the PI3K pathway, including PIKfyve, contributes to OBI efficacy (Figure S4).
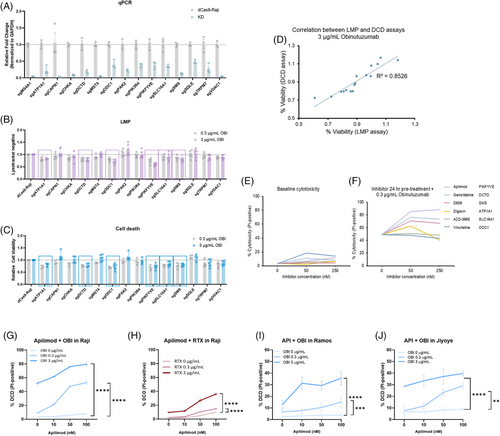
Given that PIKfyve regulates lysosomal membrane integrity,5 we investigated whether its inhibition affects lysosomal function and DCD. Apilimod, disrupts lysosomal fission and induces persistent lysosomal hypertrophy.5 Notably, the extend of OBI-induced cell death amplified by Apilimod was disproportionately greater than the increase in LMP. This pattern contrasts with the response to L-Leucyl-L-Leucine methyl ester (LLOMe), where high concentrations induced both LMP and DCD, while low concentrations triggered substantial LMP with minimal DCD (Figure 3A), likely due to activation of lysosomal membrane repair mechanisms.6 These findings suggest that OBI-induced LMP – particularly when enhanced by Apilimod – may exceed the threshold of damage that is reparable, resulting in irreversible lysosome-dependent cell death. The synergistic increase in OBI-induced LMP and DCD by Apilimod was consistently observed across multiple B-cell lymphoma cell lines, including Raji, Ramos, and Jiyoye (Figure 3A–C). To further validate LMP as the underlying mechanism, we assessed cathepsin B release as a marker of lysosomal disruption. We observed a marked increase in cytosolic cathepsin signal in cells co-treated with OBI and Apilimod compared to OBI alone (Figure 3D and E), providing direct evidence of LMP-associated protease release.
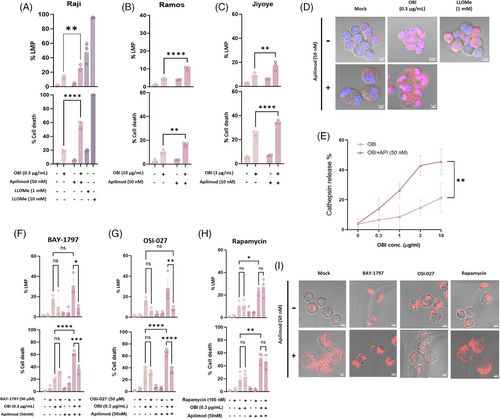
To explore the mechanisms underlying this effect, we assessed lysosomal trafficking and signalling pathways. BAY-1797, a P2X4 receptor inhibitor, significantly reduced Apilimod-enhanced DCD, suggesting that endosomal-lysosomal trafficking is essential for this process (Figure 3F). Furthermore, OSI-027, a dual mTORC1/2 inhibitor,7 completely abrogated Apilimod's effect, whereas Rapamycin had no significant impact (Figure 3G and H), our results suggest that mTORC2, rather than mTORC1, may be involved in the regulation of lysosomal homeostasis during cytotoxic stress caused by OBI. Confocal imaging confirmed that Apilimod-induced lysosomal enlargement was evident in all conditions except BAY-1797 treatment, and only OSI-027 reduced lysosome intensity in imaging (Figure 3I). These findings indicate that PIKfyve inhibition by Apilimod perturbs lysosomal homeostasis, and suggest that mTOR2 signalling and lysosomal trafficking may contribute to regulating OBI-induced cytotoxicity.
Our study identifies PIKfyve as a critical regulator of OBI-induced LMP and DCD, providing mechanistic insights into how lysosomal integrity governs B-cell lymphoma sensitivity to anti-CD20 therapy. The discovery that PIKfyve inhibition potentiates OBI cytotoxicity highlights its potential as a co-therapeutic target for lymphoma treatment. Notably, Apilimod not only enhanced OBI-induced DCD but also potentiated RTX-induced cytotoxicity, suggesting a broader role for PIKfyve inhibition in anti-CD20 monoclonal antibody therapy. The mechanistic link between PIKfyve inhibition and lysosomal dysfunction7 suggests that disrupting lysosomal integrity may be a novel strategy to enhance lymphoma therapy. Currently, PIKfyve inhibitors such as Apilimod are in clinical trials for B-cell malignancies.8 The results of this study may support further preclinical and clinical evaluation of Apilimod in combination with anti-CD20 therapies.
More precise genetic and pharmacologic analyses are needed to dissect the exact contribution of each mTOR complex to the increased OBI-induced cell death. However, the increase in OBI-induced cell death by Apilimod, which is not inhibited by rapamycin but is inhibited by OSI-027 in this study, suggests that mTORC2 rather than mTORC1 may be involved in the regulation of lysosomal homeostasis during LMP stress. However, the increase in OBI-induced apoptosis by apilimod, which is not inhibited by rapamycin but is inhibited by OSI-027, in this study suggests that mTORC2 rather than mTORC1 may be involved in the regulation of lysosomal homeostasis during LMP stress in this process. Recent studies have shown that mTORC2 prevents membrane stress-induced apoptosis by regulating lipid composition and membrane tension.9 Given that PIKfyve inhibition causes lysosomal swelling and destabilisation,10 the fact that OSI-027 inhibits OBI cell death enhanced by apilimod suggests that mTORC2 may play a potential role in maintaining lysosomal integrity under stress conditions.
Future studies should explore whether PIKfyve inhibition enhances anti-CD20 efficacy across diverse B-cell lymphoma subtypes, including diffuse large B-cell lymphoma (DLBCL). Additionally, the long-term impact of lysosomal destabilisation on lymphoma progression and immune responses warrants further investigation.
CONCLUSION
Our study establishes PIKfyve inhibition as a potent enhancer of OBI-induced DCD, uncovering lysosomal dynamics and potential involvement of mTORC2 in this process. These findings provide a strong mechanistic basis for rational combination therapies in B-cell malignancies, with implications for improving antibody-based cancer treatments
AUTHOR CONTRIBUTIONS
YK performed and analysed most of the iCas9 screening experiments, which reveal Apilimod as an OBI combination drug. JO performed both LMP and DCD-related experiments with various lysosome-regulating drugs. JRK helped the experiment using CLL cell from patient. JYK organised and supervised the whole project. YK helped and reproduced Lysotracker assays, DL supervised sgRNA library screening strategy as well as many technical processes. YK, DL, JO and JYK wrote the manuscript.
ACKNOWLEDGEMENTS
The authors are deeply grateful to Professor Jonathan Weissman (David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology) for constructing the sgRNA library. Their creation and sharing of this useful tool will serve as an important resource for the development of new drug in various fields.
CONFLICT OF INTEREST STATEMENT
All authors have declared that no competing interest exists.
FUNDING
This work was supported by grants from the National Research Foundation of Korea, Project Nos. NRF-2019R1A2C1086348 and RS-2023-00220853 to J.Y.K. This work was also supported by a faculty research grant from the Yonsei University College of Medicine (6-2022-0044) for J. Y K. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ETHICS STATEMENT
The study protocol involving human samples was approved by the Yonsei University Institutional Review Board (IRB No. 4-2022-0471), and informed consent was obtained from all participants in accordance with the Declaration of Helsinki.
CONSENT TO PARTICIPATE
Patient-derived PBMC samples were isolated with approval of Yonsei University Institutional Review Committee after obtaining informed consent, under IRP procedure (#4-2022-0471).




