Kibra knockdown inhibits the aberrant Hippo pathway, suppresses renal cyst formation and ameliorates renal fibrosis in nphp1KO mice
Yichen Yang, Zhihe Xue and Jiayong Lai contributed equally to this research.
Abstract
Introduction
Nephronophthisis (NPH) is an autosomal recessive interstitial cystic kidney disease, which is the most common genetic cause of end-stage renal disease (ESRD) in childhood. The Hippo pathway is regulated by the cilium and has been suggested to be linked to NPH. The aim of the study was to investigate the involvement of Hippo pathway in the pathogenesis of nphp1 defect-associated NPH (NPH1).
Method
Nphp1 knockout (nphp1KO) Madin-Darby Canine Kidney (MDCK) cells and nphp1KO C57BL/6J mice were generated via CRISPR gene editing strategy. The siRNAs targeting Kibra, MST1 and LATS1 were designed. An AAV9 vector was designed for Kibra knockdown. The expression and phosphorylation of core Hippo pathway molecules were evaluated. Pathological renal changes were evaluated via light microscopy respectively with haematoxylin–eosin and Masson staining.
Results
In nphp1KO MDCK cells, nphp1KO mice and NPH1 patients’ kidneys, Kibra, p-MST1/2, p-LATS and p-YAP exhibited a notable increase in levels, with an even greater elevation observed in renal cyst cells, indicating the Hippo pathway activated in these nphp1-deficient contexts. Nphp1 re-expression reversed the Hippo pathway activation in cells, indicating that the Hippo pathway activation is related to nphp1 deficiency in vitro. Meanwhile, in vitro, MST1 knockdown downregulated LATS1 and YAP phosphorylation, LATS1 knockdown downregulated YAP phosphorylation, suggesting the activation of the canonical Hippo pathway in nphp1-deficient contexts. Knockdown of the upstream regulator Kibra inhibited the Hippo pathway activation in both nphp1KO MDCK cells and mice. Following Kibra knockdown, the organisation of nphp1KO MDCK cells became more compact, the intensity of the actin fibres increased. Besides, decreased renal fibrosis and cyst formation were observed in nphp1KO mice.
Conclusions
The canonical Hippo pathway is aberrantly activated in nphp1-deficient conditions. Kibra may serve as a crucial upstream regulator of nphp1 deficiency-related Hippo pathway activation. Kibra upregulation and activation of the Hippo pathway are involved in the pathogenesis of NPH1.
Key Points
- Canonical Hippo pathway activated in nphp1-deficient disease models and patients.
- Kibra was a key upstream molecule in regulating the activation of canonical Hippo pathway in nphp1-deficient disease models and patients and closely related to renal cyst formation and fibrosis in nphp1KO mice.
1 INTRODUCTION
Nephronophthisis (NPH) is an autosomal recessive interstitial cystic kidney disease that accounts for 10%–20% of the end-stage renal disease (ESRD) cases in children and adolescents.1 Over 20 causative genes have been identified, among which nphp1 is predominant. Nphp1 mutation causes type 1 NPH (NPH1).2-4 Typical pathological changes in patients with NPH include corticomedullary tubular atrophy, dilatation, cyst formation, tubular basement membrane thickening and layering, and diffuse tubulointerstitial fibrosis with inflammatory cell infiltration.5, 6 The pathogenesis of NPH is unclear and no effective therapy has been developed for its clinical management. The proteins encoded by pathogenic genes associated with NPH are structural components of primary cilia and/or are associated with ciliary function.7 Primary cilium is antenna-like sensory organelles that protrude from the surface of most vertebrate cells.8 The primary cilium extends from the basal body as a solitary unit, detects extracellular changes and transduces signals intracellularly via different signalling pathways: Hippo, Wnt and TGF-β pathways,9 to regulate cellular activity, development and homeostasis. Ciliary defects caused by genetic defects cause a group of diseases known as ciliopathies, including autosomal dominant/recessive polycystic kidney disease (ADPKD/ARPKD).
The Hippo pathway, which was identified approximately 30 years ago during tissue growth screening in Drosophila melanogaster, is an evolutionarily conserved signalling network and is considered as a key player in a number of cellular functions, including cell proliferation, apoptosis, differentiation, organ size regulation, tumourigenesis.10-13 The Hippo pathway comprises three groups of components: upstream regulators, core molecule proteins and downstream effectors. In mammals, the core components include mammalian sterile 20-like kinases 1 and 2 (MST1/2), salvador homolog 1 (SAV1), large tumour suppressor 1 and 2 (LATS1/2) and Mps one Binder (MOB) kinase activator 1A and 1B (MOB1A/1B). Yes-associated protein (YAP) and transcriptional coactivator with PSD-95/discs large/ZO-1 (PDZ) -binding motif (TAZ) are the main downstream effectors. When the Hippo pathway is activated, MST1/2 interact with SAV1 and then phosphorylate SAV1, MOB1 and LATS1/2.14-16 LATS1/2 then phosphorylate YAP and TAZ directly, phosphorylated YAP and TAZ bind to cytoplasmic 14-3-3 proteins and are then retained in the cytoplasm or degraded by ubiquitylation.17-20 When the Hippo pathway is inactivated, dephosphorylated YAP and TAZ translocate into the nucleus and bind to TFs, mainly TEA domain transcription factor 1–4 (TEAD1-4), to promote or, infrequently, inhibit target gene transcription.21 Proteins involved in cell polarity and cell adhesion are upstream regulators of the Hippo pathway. Among these proteins, the most important are kidney and brain expressed protein (Kibra; also known as WWC1), Merlin (Mer; NF2 in mammals) and Expanded (Ex; FRMD6 in mammals). Kibra is a scaffolding protein with two WW domains, a C2-like domain and a glutamic acid-rich C-terminus. It is predominantly expressed in the kidney and brain tissues and acts with NF2 and FRMD6 as an upstream regulator of the Hippo pathway.22, 23 Kibra can directly bind to LATS1/2 and SAV1, bringing LATS1/2 in proximity to the MST1/2-SAV1 complex and promoting the phosphorylation of LATS1/2 by MST1/2.24
YAP deletion is lethal.25 Conditional knockout of YAP in the nephrogenic lineage causes severe nephrogenesis defects.26 TAZ deletion causes cystic kidney disease in mice and renal cysts in zebrafish. Renal cyst formation at the corticomedullary junction mimics NPH. A panel of pathogenic genes associated with ciliopathies, such as Dctn5, Kif3a, Pkhd1, Tsc1 and Ofd1, were downregulated upon TAZ deletion.27, 28 Previous studies have demonstrated that mutations in nphp4,29 nphp9/Nek8,30 nphp331 and nphp16/Anks632 can modify the activity of the Hippo pathway through direct or indirect interactions with YAP or TAZ. However, the role of the Hippo pathway in the pathogenesis of NPH remains largely unexplored.
In this study, the association of the Hippo pathway with NPH1 was explored using a nphp1 knockout (nphp1KO) mouse model, nphp1KO MDCK cells and renal biopsy specimens from NPH1 patients. We found that the canonical Hippo pathway was aberrantly activated, with sequential activation of the core molecules MST1/2 and LATS1/2, and the effectors YAP/TAZ. The upstream regulator Kibra was found to be significantly upregulated. Kibra knockdown partially reversed the activation of the Hippo pathway in nphp1-deficient models both in vitro and in vivo, suppressed renal cyst formation and ameliorated renal fibrosis in nphp1KO mice.
2 MATERIALS AND METHODS
2.1 Cell culture
The wild-type (WT) Madin-Darby Canine Kidney (MDCK) cell line was kindly provided by Professor Xueqing Yu (Kidney Institute, Sun Yat-sen University, China). The nphp1-knockout (nphp1KO) MDCK cell line was generated previously by our group using CRISPR-Cas9 gene editing, firstly.33, 34 To get a more stable cell line, we ultimately utilised the nphp1KO MDCK cell line constructed by Ubigene Biosciences through CRISPR gene editing strategy. Briefly, the knockout was achieved by targeting exons 3–6 of the nphp1 gene. Plasmids expressing Cas9 protein and gRNAs were introduced into MDCK cells via electroporation. Single-cell clones were selected and validated through polymerase chain reaction (PCR) and genomic sequencing. All the cells were cultured in minimal essential medium (Gibco) supplemented with 10% foetal bovine serum (FBS; Gibco) and 1% sodium pyruvate (Gibco).
2.2 Mice
nphp1KO C57BL/6J mice were generated by our group via deletion of exons 2–20 of nphp1 using CRISPR-Cas9 gene editing.35, 36 (AAV9 vectors carrying either recombinant Kibra-cDNA plasmids or control plasmids were generated by GeneChem Biotechnology. These vectors were administered via tail vein injection to 4-week-old mice, with each mouse receiving 8 × 1010 copies. Each experimental group consisted of six mice. All mice were euthanised at 12 weeks of age using sodium pentobarbital anaesthesia. All animal experiments were performed according to the Animal Research: Reporting of In Vivo Experiments (ARRIVE 1) guidelines and the protocols were approved by the Institutional Animal Care and Use Committee of Nanfang Hospital, Southern Medical University (Application Number: NFYY-2020-0736).
2.3 Patient renal tissue
Renal tissue samples from three NPH1 patients and three control patients were obtained via renal biopsy. The three NPH1 patients were 13, 13 and 10 years old, respectively. All harboured the same mutation: homozygous large deletion of nphp1. Three control patients were ailing with primary nephrotic syndrome: Control patient 1 was a 14-year-old male with minimal change kidney disease. Control patient 2 was an 11-year-old male also with minimal change kidney disease. Control patient 3 was a 10-year-old female with focal segmental glomerulosclerosis (FSGS). This study was reviewed and approved by the Ethics Committee of Nanfang Hospital (NFEC-2019-047) of Southern Medical University.
2.4 Transfection
LATS1, Kibra, MST1 and negative control small interfering RNAs (siRNAs) were constructed by HANYI. The siRNA sequences used were as follows: LATS1 siRNA, 5′-GAUAAAGACACUAGGAAUATT-3′; Kibra siRNA, 5′- CCCGGAAGCGGUUGGAGAATT-3′; and MST1 siRNA, 5′- CAGAAGUGAUUCAGGAAAUTT-3′. pcDNA3.1-nphp1 was synthesised by GenePharma. GP-transfect-Mate transfection reagent (GenePharma) was used for transfection, as instructed by the manufacturer. Six hours after transfection, the FBS-free medium was replaced with medium containing 10% FBS. Subsequently, the cells were cultured for 24 h before RNA extraction or for 48 h before protein extraction.
2.5 RNA extraction and real-time quantitative PCR (qPCR)
Total RNA was extracted using RNA Isolator Total RNA Extraction Reagent (Vazyme) and reversed using HiScript II Q RT SuperMix (Vazyme) according to the manufacturer's instructions. ChamQ SYBR qPCR Master Mix (Low ROX Premixed; Vazyme) was mixed with the primers and cDNA, and a QuantStudio™ 5 Real-Time PCR System (Thermo Fisher) was used to perform qPCR. The relative mRNA expression levels were calculated using the 2−ΔCt method. The primer sequences are shown in Supporting Information Table 1.
2.6 Protein extraction and western blotting
Cells and tissues were lysed via Radio Immunoprecipitation Assay Lysis buffer (RIPA Lysis buffer; Beyotime) supplemented with 1% Phenylmethylsufonyl fluoride (PMSF; Fudebio) and 1% phosphatase inhibitor (8Solarbio). The lysates were clarified by sonication and centrifugation. Subsequently, the diluted proteins were heated at 100°C for 10 min. Western blotting was performed following standard protocols. These blots were incubated with primary antibodies (Supporting Information Table 2) and then with secondary antibodies (Supporting Information Table 2). The blots were visualised with a Tanon scanner. The density of each band was normalised to that of the GAPDH band using Image J software. And three different biological replicates were used for statistical analysis.
2.7 Immunohistochemistry (IHC), haematoxylin–eosin (H&E) staining and Masson's trichrome (Masson) staining
Paraffin-embedded kidney sections were sliced into a 4 µm thickness and used after dewaxing and rehydration. For IHC, antigen retrieval was performed in target retrieval solution using microwave oven. The activity of endogenous peroxidase was blocked using 3% H2O2. The sections were then blocked with a 5% bovine serum albumin (BSA) solution for 1 h, followed by incubation with primary antibodies (Supporting Information Table 2) at 4°C overnight. A universal two-step assay kit (PV-9000; ZSGB-BIO) and a DAB kit (ZLI-9018; ZSGB-BIO) were used for staining. An upright microscope (ECLIPSE Ci-L plus, Nikon) or an SQS40P Slide Scan System (Shengqiang Technology) was used to examine the sections. For H&E and Masson staining, eosin (DH0055; Leagene) and a Masson's Trichrome Stain Kit (G1340; Solarbio), respectively, were used following manufacturer's protocol. All images were analysed by the ImageJ software.
2.8 Statistical analysis
Data are presented as the mean ± standard error of the mean (SEM). A p value <.05 was considered statistically significant. Data were analysed using GraphPad Prism version 8.0.2.
3 RESULTS
3.1 Nphp1 deficiency activates the canonical Hippo pathway in MDCK cells
To explore the status of the Hippo pathway in the context of NPH1, nphp1KO MDCK cells were generated by deleting exons 3–6 of nphp1 using CRISPR-Cas9 gene editing. qPCR and western blot results demonstrated a significant reduction in nephrocystin-1 expression in nphp1KO MDCK cells (Figure 1A–C). The total expression levels of the key components of the Hippo pathway were measured. The protein levels of these key components remained unchanged (Figure 1D–H). However, the phosphorylation of MST1/2, LATS1, YAP and TAZ were significantly increased (Figure 1D–H), indicating that the Hippo pathway was activated in nphp1KO MDCK cells.
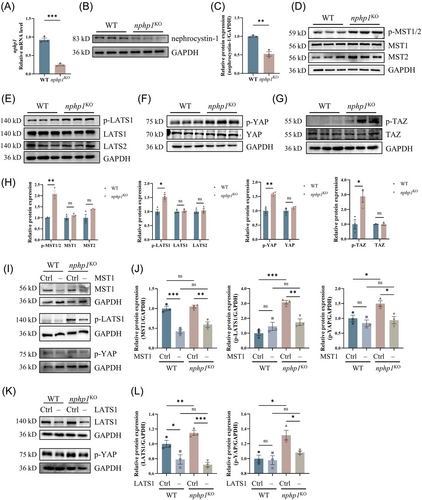
The Hippo pathway can be activated via canonical and non-canonical mechanisms. To determine which pathway was dominantly activated in nphp1KO MDCK cells, we generated siRNAs targeting MST1 and LATS1 and transfected them into both WT and nphp1KO MDCK cells, respectively (Figure 1M). As shown in Figure 1I,K, the expression of MST1 or LATS1 was successfully knocked down. MST1 knockdown significantly decreased both p-LATS1 and p-YAP levels (Figure 1I,K), while LATS1 knockdown significantly decreased p-YAP level in nphp1KO MDCK cells (Figure 1K,L). These results indicated that the Hippo pathway is activated through the canonical mechanism in nphp1KO MDCK cells.
3.2 Nphp1 re-expression reversed the Hippo pathway activation in nphp1KO MDCK cells
To verify whether the activation of the Hippo pathway in the NPH1 model resulted from nphp1 deficiency, we constructed a nphp1 overexpression lentiviral plasmid and transduced it into nphp1KO MDCK cells. Successful re-expression of nphp1 was confirmed by western blotting (Figure 2A,B). Re-expression of nphp1 in nphp1KO MDCK cells led to a reversal in the increased levels of p-MST1/2, p-LATS1 and p-YAP, whereas no significant alterations were observed in the levels of the non-phosphorylated forms of the core Hippo pathway proteins (Figure 2C–F and Supporting Information Figure 1). These findings indicated that nphp1 re-expression reversed the abnormal activation of the Hippo pathway in nphp1KO MDCK cells and that the activation of the Hippo pathway was associated with the deficiency of nphp1 in nphp1KO MDCK cells.
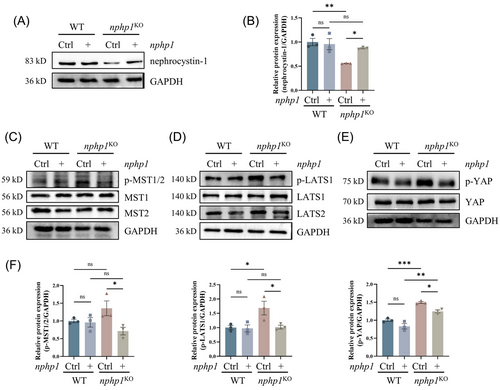
3.3 The Hippo pathway is activated in the kidneys of nphp1KO mice and human NPH1 patients
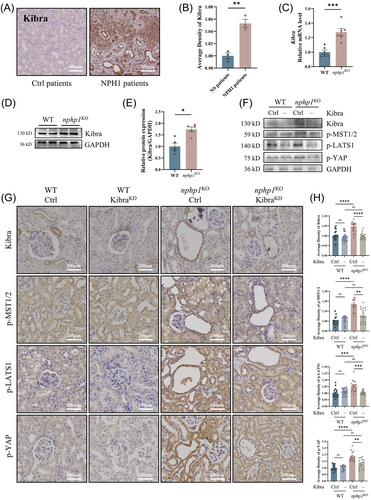
To assess if the Hippo pathway is also activated in vivo, we used nphp1KO mice with an NPH renal phenotype, previously generated by our group using CRISPR-Cas9 gene editing.36 The knockout efficiency of nephrocystin-1 in nphp1KO mice was reconfirmed by western blotting (Figure 3A,B). As expected, p-MST1/2, p-LATS1 and p-YAP levels in renal tissue exhibited an increase in nphp1KO mice, while the levels of non-phosphorylated LATS1, LATS2, MST1, MST2 and YAP were not significantly changed in nphp1KO mice compared with WT mice (Figure 3C–E). IHC revealed strong positivity for p-LATS1 in nphp1KO mice's renal cyst cells, but weak positivity in WT mice (Figure 3G).
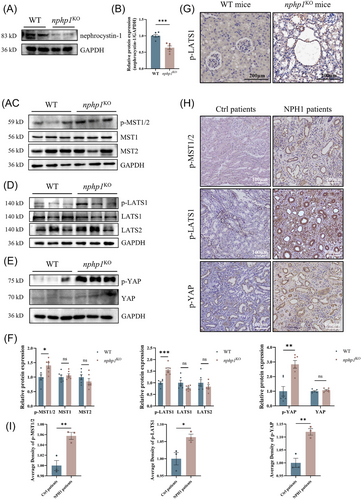
Furthermore, we selected 3 NPH1 patients and 3 control patients with primary nephrotic syndrome and found that phosphorylated MST1/2, LATS1 and YAP levels in renal tissue were higher in NPH1 patients than in nephrotic syndrome patients (Figure 3H). In the renal cysts of NPH1 patients, the phosphorylation of these molecules was more prominent (Figure 3I).
These results suggested that the Hippo pathway was also activated in the kidneys of both nphp1KO mice and NPH1 patients and was much more strongly activated within renal cyst cells, highlighting its potential role in renal cyst formation in NPH.
3.4 Kibra knockdown inhibits the activation of the Hippo pathway in nphp1KO MDCK cells
Despite the abovementioned findings, the mechanism through which nphp1 deficiency induces the activation of the Hippo pathway is not clear. The canonical Hippo pathway activation is related to a set of upstream activation factors such as Kibra, NF2 and FMRD6. Next, we detected the expression of the molecules upstream of the Hippo pathway in nphp1KO MDCK cells. Compared with the expression levels in WT MDCK cells, Kibra and FMRD6 mRNA expression was upregulated in nphp1KO MDCK cells, with Kibra upregulation being more significant. The change in NF2 expression was not statistically significant (Figure 4A). Western blotting analysis confirmed the upregulation of Kibra protein expression (Figure 4B,C).
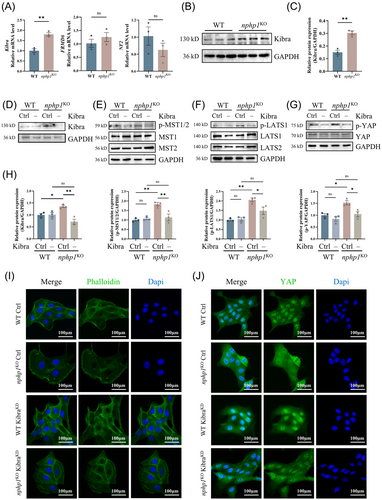
To explore whether Kibra upregulation mediates the activation of the Hippo pathway in nphp1KO models, we transfected both WT and nphp1KO MDCK cells with Kibra knockdown siRNA (si-Kibra) and control siRNA (si-Ctrl), respectively. The efficiency of Kibra knockdown was confirmed by western blotting (Figure 4D,H). Although there were no significant changes in the levels of MST1/2, LATS1/2 and YAP, the phosphorylation of MST1/2, LATS1 and YAP decreased significantly in nphp1KO MDCK cells upon si-Kibra transfection compared to si-Ctrl transfection (Figure 4E–H). In the WT group, the changes in the levels of both phosphorylated and non-phosphorylated forms of these proteins were not significant (Figure 4E–H).
Besides, WT MDCK cells were tightly arranged with clear outlines, and their actin fibres were organised around the periphery of the cell. However, in nphp1KO MDCK cells, the arrangement of cells was disturbed with increased volume in a few cells. The actin fibres surrounding these enlarged cells exhibited weaker intensity. Upon knockdown of Kibra, the organisation of nphp1KO MDCK cells became more compact, and the intensity of the actin fibres increased (Figure 4I).
As we expected, in WT MDCK cells, YAP was distributed in both cytoplasm and nucleus, with evident nuclear localisation. In nphp1KO MDCK cells, however, YAP was more inclined to be distributed in the cytoplasm. Knocking down Kibra promoted nuclear localisation of YAP in nphp1KO MDCK cells (Figure 4J and the statistical analysis is presented in Supporting Information Figure 2).
3.5 Kibra knockdown inhibits the activation of the Hippo pathway in nphp1KO mice
We further evaluated the expression of Kibra in vivo. Kibra was upregulated in the renal tissues of NPH1 patients (Figure 5A,B) and in the kidneys of nphp1KO mice (Figure 5C–E). To explore the effect of Kibra knockdown on the Hippo pathway activation in nphp1KO mice, we designed a Kibra knockdown short hairpin RNA (shRNA) construct and inserted it into an AAV9 vector (KibraKD-AAV9). We introduced KibraKD-AAV9 into both WT and nphp1KO mice at 4 weeks of age via tail vein injection. The efficiency of Kibra knockdown in the kidney tissue was confirmed by western blotting (Figure 5F) and IHC (Figure 5G,H). Upon Kibra knockdown, the phosphorylation of MST1/2, LATS1 and YAP decreased in the kidneys of nphp1KO mice, but the differences in the phosphorylation of these proteins between the kidneys of WT mice without or with Kibra knockdown were not significant (Figure 5F–H and Supporting Information Figure 3). Moreover, the Kibra, p-MST1/2, p-LATS1 and p-YAP were more positive in or around the renal cysts. These results suggested that Kibra knockdown can inhibit the activation of the Hippo pathway in the kidneys of nphp1KO mice.
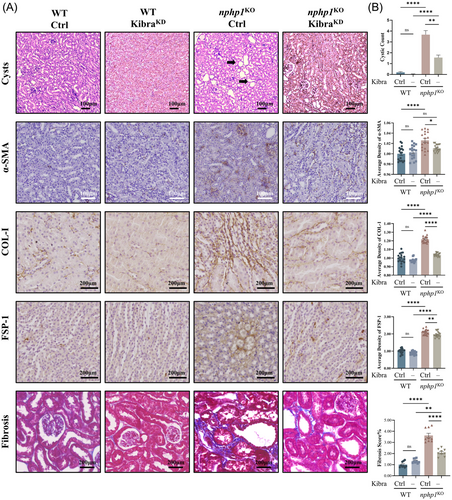
3.6 Kibra knockdown suppresses renal cyst formation and ameliorates renal fibrosis in nphp1KO mice
Our pathological analysis revealed a significant decrease in renal cyst formation in Kibra-knockdown nphp1KO mice compared to that in negative control nphp1KO mice (Figure 6A,B). IHC showed that expression of α-SMA, COL-1, FSP-1 was lower in Kibra-knockdown nphp1KO mice than in negative control mice (Figure 6A,B). The renal fibrosis score was evaluated by Masson staining, which also revealed an improvement in renal fibrosis in Kibra-knockdown nphp1KO mice compared to negative control mice (Figure 6A,B). These findings reveal that Kibra knockdown can effectively reduce renal cyst formation and ameliorate fibrosis in nphp1KO mice, suggesting that Kibra may serve as a potential therapeutic target for NPH1.
4 DISCUSSION
In this study, by using nphp1 knockout models, we demonstrated that the canonical Hippo pathway was aberrantly activated in the context of nphp1 deficiency both in vitro and in vivo and was highly activated in nphp1-deficient renal cyst cells. nphp1 deficiency induced the activation of the canonical Hippo pathway via the sequential phosphorylation of all the core molecules. When nphp1 was re-expressed in nphp1KO MDCK cells, the activation of the Hippo pathway was inhibited. Activation of the Hippo pathway has previously been observed in the context of defects in other NPH-associated pathogenic genes, such as nphp4, nphp9/Nek8, nphp3 and nphp16/Anks6. Nephrocystin-4 inhibits phosphorylation of YAP and TAZ via direct interaction with LATS1. This inhibition is removed when nphp4 is knocked down, leading to the phosphorylation and cytoplasmic sequestration of YAP.29 Nek8 directly interacts with TAZ to form the TAZ/Nek8 complex, which facilitates the nuclear translocation of TAZ.30 Nephrocystin-3 promotes TAZ/TEAD-driven transcriptional activity.31 Anks6 mutants have been found to exhibit altered ciliary localisation, promote YAP expression and increase both its nuclear and cytoplasmic accumulation.37 We found that the upstream molecule of the Hippo pathway, Kibra, was prominently upregulated in the context of nphp1 deficiency. Kibra knockdown decreased the phosphorylation of the core proteins MST1/2 and LATS1 as well as the phosphorylation of YAP in nphp1KO models. Our results showed that the canonical Hippo pathway was activated in models of nphp1 deficiency via the sequential activation of core proteins. Furthermore, Kibra knockdown reduced renal cyst formation and ameliorated renal fibrosis in nphp1KO mice.
The Hippo pathway controls organ size and regeneration by regulating cell proliferation, differentiation, survival and apoptosis. In ADPDK, inactivation of the Hippo pathway has been evidenced and may facilitate continuous growth of renal cysts until the late stage of the disease, leading to the enlargement of kidneys and the loss of their normal shape.38 In contrast, the Hippo pathway is active under most conditions in NPH, as reported previously29, 30, 32 and observed in this study in nphp1KO models and NPH1 patients. Activation of the Hippo pathway in NPH has been proposed to limit cyst growth and contribute to maintaining the normal size of the kidney.38 We observed a significant enhancement in the levels of p-MST1/2, p-LATS and p-YAP within renal cyst cells in NPH1 patients, which may indeed limit cyst growth and contribute to maintenance of the kidney size. However, when the Hippo pathway was inhibited by Kibra knockdown, the number of renal cysts decreased in the nphp1KO mice, indicating that the mechanism of renal cyst formation in NPH may be far more complex than previously thought and is worthy of further study.
We also observed that Kibra knockdown and Hippo inhibition ameliorated renal fibrosis in nphp1KO mice. In contrast to the situation in ADPKD, in which renal fibrosis develops only at a late stage and may be secondary to renal cyst growth, renal fibrosis in NPH develops at an early stage, even before renal tubular dilatation and/or cyst formation.39, 40 Therefore, renal fibrosis in NPH is considered to be an intrinsic disorder, although the underlying mechanism is unclear.
TGF-β/SMAD and Wnt pathways are the two main pathways that mediate renal fibrosis in different contexts of kidney impairment. Both signalling pathways are associated with cilium.9 Many studies have shown that Hippo pathway participates in crosstalk with both TGF-β and Wnt signalling pathways at different levels.41 However, these crosstalks are highly context-specific, and their association with renal fibrosis in NPH still needs to be determined.
In addition to its role as a key upstream regulator of the Hippo pathway in regulating cell proliferation, Kibra could regulate the actin cytoskeleton and focal adhesions either directly or indirectly. Kibra associates with synaptopodin directly to regulate the actin cytoskeleton in podocytes.42 Kibra can directly interact with the scaffolding protein angiomotin (Amot) to enhance the assembly of actin filament at focal adhesions in endothelial cells.43, 44 Kibra overexpression disrupts the actin cytoskeleton structure in podocytes.45, 46 Podocyte-specific Kibra overexpression in vivo predisposed experimental mice to acute and chronic podocyte and glomerular impairment.46 Increased Kibra expression was observed in patients' cohorts of FSGS and glomerular kidney disease, where it was found to be associated with glomerular disease progression.45, 46 Consistent with these findings, we also found that upregulation of Kibra participates in the disarrangement of the actin skeleton in renal tubular cells in the context of nphp1 deficiency. Kibra expression was increased in renal tubular cells in both the in vivo and in vitro nphp1KO models, as well as in renal biopsy tissue from NPH1 patients. Changes in cell morphology and actin cytoskeleton disarrangement were observed in nphp1KO MDCK cells. Kibra knockdown reversed the alterations in cell morphology and disarrangement of the cytoskeleton in nphp1KO MDCK cells. Whether the disarrangement of the actin cytoskeleton associated with Kibra upregulation contributes to disease progression and renal fibrosis in NPH1 deserves further investigation. These findings indicated that Kibra may be a potential therapeutic target for NPH1.
Overall, our results showed that the canonical Hippo pathway was aberrantly activated both in vivo and in vitro models of nphp1 deficiency, as well as in the kidneys of human NPH1 patients, and was highly active in renal cyst cells. Activation of the Hippo pathway may contribute to the maintenance of kidney size and the inhibition of renal cyst growth in patients with NPH. Kibra, a key upstream regulator of the Hippo pathway, was significantly upregulated in the context of nphp1 deficiency. Kibra upregulation was associated with aberrant activation of the Hippo pathway and cytoskeletal disarrangement, as well as with renal cyst formation and renal fibrosis. However, further studies are needed to clarify the mechanism by which the Kibra and the Hippo pathway contribute to the pathogenesis of NPH1.
AUTHOR CONTRIBUTIONS
Conceptualisation: Liangzhong Sun. Data curation: Yichen Yang, Zhihe Xue, Jiayong Lai. Formal analysis: Yichen Yang, Zhihe Xue, Jiayong Lai. Funding acquisition: Liangzhong Sun, Yaqing Liu. Investigation: Yichen Yang, Zhihe Xue, Jiayong Lai, Jinglan Zhang, Changmiao Pang, Jinglin Zhong, Zhanpeng Kuang, Baojuan Zou. Project administration: Liangzhong Sun. Resources: Liangzhong Sun, Yaqing Liu. Supervision: Liangzhong Sun, Yaqing Liu. Validation: Yichen Yang, Zhihe Xue, Jiayong Lai, Jinglan Zhang. Visualisation: Yichen Yang, Zhihe Xue, Jiayong Lai. Writing—original draft: Yichen Yang, Zhihe Xue, Jiayong Lai. Writing – review & editing: Liangzhong Sun, Yaqing Liu.
ACKNOWLEDGEMENTS
We are thankful to Albert CM Ong (Kidney Genetics Group, Academic Unit of Nephrology, University of Sheffield Medical School, UK) for his kind suggestions for this research. This study was supported by the National Natural Science Foundation of China (Grant No. 81670610). This work was partially supported by the Natural Science Foundation of Guangdong Province (Grant Nos. 2022A1515012307 and 2024A1515013163).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in this paper and the supporting information of this article.
The use of clinical samples for the present study was approved by the Ethics Committee of Nanfang Hospital (NFEC-2019-047) of Southern Medical University. The animal experiments were approved by the Institutional Animal Care and Use Committee of Nanfang Hospital, Southern Medical University (Application Number: NFYY-2020-0736).




