Integrative analysis identifies three molecular subsets in ovarian cancer
Bo Liu is the first author
Dear Editor,
We identified three novel molecular subtypes (tumour-enriched, mixed and immune-enriched) of ovarian cancer (OC) and revealed a complex molecular landscape among three subtypes. Several researches have studied the molecular subtypes of OC,1 yet more well-recognized subtypes and more comprehensive analysis are needed. In our study, a multi-platform analysis was implemented.
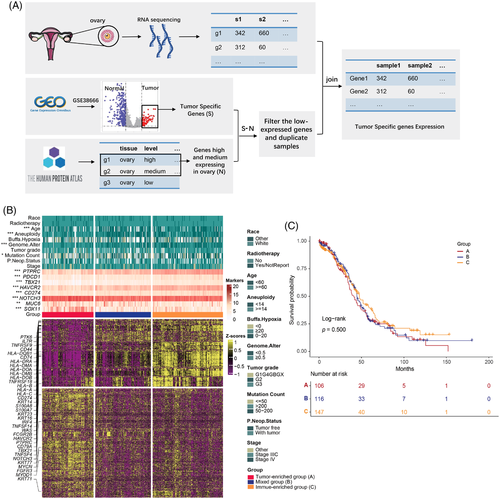
To profile the global genomic patterns of OC, a total of 4,188 tumour-specific genes (Figure S1) were screened in 376 patients from The Cancer Genome Atlas, Gene Expression Omnibus and The Human Protein Atlas datasets (Figure 1A). Then, an unsupervised hierarchical clustering analysis was performed to reveal three subgroups in these OC patients (Figure 1B). Analysis of the canonical markers for lineage-specific different expression genes (DEGs) suggested different tumour cell compositions among the three subgroups. OC patients of cluster A showed the highest expression of OC-associated oncogenes KRT16 and KRT23,2 and the lowest expression of immune cell markers PTPRC and PDCD1, suggesting enrichment of tumor cells. While OC samples of cluster C displayed the highest expression of the immune cell markers PTPRC, PDCD1, HAVCR2 and CD2743 and exhibited low or no expression of the cancer-related oncogenes (TBX21 and NOTCH34), indicating predominance of immune cells. And cluster B presented mixed expression characteristics of both cancer and immune cells, and was therefore classified as ‘mixed group’. We further examined the clinical factors of genomics-based classification. Cluster A had significantly higher aneuploidy score and genome altered fraction than cluster C (p < .001), but cluster A and B had significantly lower diagnosis age and mutation count (p < .05, Figure 1B and Tables S1 and S2). Additionally, three groups have similarities on race, figo-stage and hypoxia-score. We also observed a trend of shortened survival in cluster A patients, although it did not reach a statistical significance in this study (log-rank p = .500, Figure 1C, Figure S2 and Table S3). These results suggested similarities and differences in clinical factors, which can help with diagnosis and treatment.
Pathways enrichment analysis was performed in cluster A to explore the dysregulated molecular processes informed by the genomic data. A total of six up-regulated KEGG: Kyoto Encyclopedia of Genes and Genomes pathways were screened, which mainly included oncogenic signallings (Figure 2A) such as WNT and MAPK signaling pathway, highlighting the importance of targeting the WNT and MAPK pathway as interested therapeutic strategies. Next, 777 genes highly abundant in cluster A were selected (Figure 2B), 42 of which were identified as functionally important (Figure 2C), which included 27 cancer testis antigens (CTAs) such as TPPP2, TAF7L and PRM1 etc., TNFSF4 as an immunotherapy candidate gene involved in promising immunological response in carcinomas,5 four transcription factors (TFs) including NEK5,6 MYT1, FGFR3 and BRSK2 as known altered genes in numerous cancer types, four genes involved in the metabolic process of glycolysis, and four enzymes correlated with metastasis (Figure 2C). A large portion of these genes were important targets of CTAs. Generally, DEGs in cluster A predominantly exhibited oncogenic function, some of which were druggable targets, which could provide effective therapies in molecular aspects.
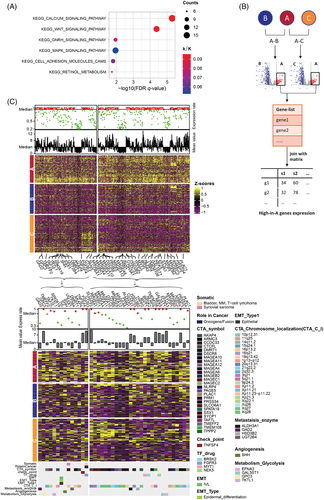
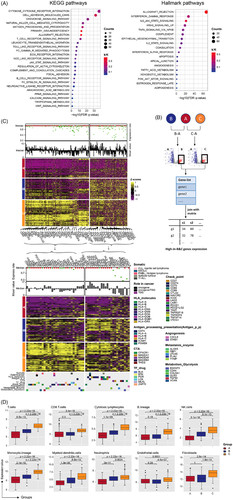
Using similar methods, we performed analysis in OC samples of cluster B and C. A total of 43 up-regulated Hallmark (n = 18) and KEGG (n = 25) pathways were identified as predominant immune-related pathways (Figure 3A). And we completed a stepwise filtering process to obtain 530 genes with high expression in cluster B and C (Figure 3B), 53 of which were annotated as functionally important immune-related genes (Figure 3C). Among of them, 17 genes (CD70, LAG3, TIGIT, etc.) were approved to be immune checkpoints, which could regulate the immune system.7 And nine genes belonged to HLA molecules, and another five genes were targeted genes for CTAs. Also, some genes like CD74,8 ICAM1 and PSMB9 were enrolled into the process of antigen processing and presentation, and some other genes were associated with TF correlated drugs, metabolic enzymes promoting cancer-cell migration, and metabolic glycolysis promoting tumorigenesis. Meanwhile, MCP-counter9 was applied to produce the absolute abundance scores of eight immune cells and two stromal cells in tumour samples (Figure 3D), which were significantly higher in cluster B and C. Together, our results indicated that cancer cells might reprogram OC's tumour microenvironment to promote cancer development, and immune checkpoint inhibitors may produce an effective treatment for subtype C.
Then, we performed whole-exome sequencing analysis to filter out the most frequently altered genes (n = 35) on OC samples (n = 250). The top three genes sorted by mutation frequencies were TP53, MYC and NDRG1, among of which, TP53 had a high mutation rate up to 95%, and the other two were with high amplification counts. And the amplification rate of MYC, NDRG1, EIF3E and SMARCA2 was significantly higher in cluster B and C, but NOTCH3 and AKT2 for cluster A (p < .05, Figure 4A and Table S4). DNA mutation sites with missense mutation or truncating and cancer hotspots of four genes including TP53, KMT2C, CDK12 and BRCA2 were presented across three subgroups of OC patients (Figure 4B and Tables S5–S8). Then, we observed the performance of copy number alteration in the three groups (Figure 4C) and discovered that segment amplification of genes such as CD27, KRAS and PTK6 were significantly more occurrence in group A on the whole, but TP53 was with significantly more deletion in group B (Figure S3). All the integrated mutational profiles supported the genomics-based clustering designation.

Results of survival analysis and pathways enrichment revealed survival differences between Figo-stage subgroups (≤ IIIB vs. ≥ IIIC) in immune-enriched group, which displayed that this new classification could augment the prognostic power as an independent clinical factor (Figure S4 and Table S9). Additionally, performance of DNA methylation and protein expression among three groups was also correlated with the genomics-based classification (Figure S5). Comparison with previous subtypes indicated the consistency in molecules and experiments with two datasets validated our clustering (Figure S6).
CONCLUSION
Novel molecular subtypes of OC was identified with molecular heterogeneity and was useful for targeted therapy. Tumor-enriched A can use targeted-tumor approach while immunotherapy is suitable for group C, and group B can use a mixture of tumor killing and immunotherapy. The complex molecular landscapes of subtypes may help with precision medicine.
ACKNOWLEDGEMENTS
The authors appreciate the TCGA and GEO database for their help with the public data enrolled in this study.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
FUNDING INFORMATION
National Natural Science Foundation of China, Grant Number: 62076015.




