Nuclear circRNA: Impairing genome stability via circR loops
Abstract
Conn et al. have recently reported circRNA: DNA hybrids (circR loops) to promote oncogenic gene translocations and impair genome stability which drive leukemia. Herein, we will summarize potential molecular mechanisms and discuss clinical significance of uncovering circR loops as endogenous RNA carcinogens.
Circular RNAs (circRNAs) are covalently closed circles of single-stranded RNA produced from non-canonical, back-splicing of pre-mRNA, which were identified more than 40 years ago.1 Compared with linear mRNAs, circRNAs exhibit structure stability following Ribonuclease R (RNase R) digestion due to the lack of classic 3′ and 5′ ends recognized by RNase R. With the fast development of RNA-sequencing over the last decade, thousands of circular RNAs in eukaryotes have been detected, and some of them even present greater expressions than linear mRNAs. The distribution of circRNAs in human bodies is organ-specific and mainly concentrated in brain.2 Various types of circRNAs have been confirmed to be related to disease progress such as cancers, diabetes, neuropsychiatric disorders and cardiovascular diseases.
Investigating molecular mechanisms by which circRNAs regulate disease progress is of great importance. In the previous studies, most characterized circRNAs, especially exonic circRNAs, were dominantly localized in the cytoplasm and verified with the competence to affect gene expression and protein function via multiple types of non-coding or coding patterns.3 CircRNAs exerting the molecular functions in the nucleus were partly similar with those patterns in the cytoplasm such as forming sponges for proteins. But differently, nuclear circRNAs could directly modulate transcription and splicing by generating circRNA: DNA hybrids (circR loops), which presented a stronger bond strength than the linear RNA: DNA hybrids.4 CircR loops have been revealed to result in transcriptional pausing5; however, it still remains elusive about underlying molecular mechanisms and biological significance.
To our excites, Conn and the colleagues recently have demonstrated four effects caused by circR loops jointly leading to oncogenic translocations and consequently driving leukemia.6 The four effects include: (1) pausing RNA polymerase II (RNA Pol II); (2) promoting DNA double-stranded breaks (DSBs); (3) altering chromosome organization; and (4) inhibiting the proteasome. Inhibiting the proteasome, which is known to be needed for stabilizing RNA Pol II7 and repairing DSBs,8 may account for the dysfunction of RNA Pol II and increase in DSBs.
In specific, the authors found that the proportion of circR loops in all the detected R loops was approximately 6%. Both circR loops and circRNA were RNase R-resistant. Over-representation enrichment analysis (OEA) on the circR loops gene set from HEK293T cells indicated that circR loops were enriched in leukemias and chromosomal changes. Because at the beginning stage of acute leukemias, formation of a potent fused oncogene by chromosomal translocations between the mixed lineage leukemia (MLL) gene and partner genes (e.g., MLLT1) was an initiating factor and could be observed in more than 70% of infant leukemias,9 the authors constructed different circMLLs in which circMLL(9,10) (hsa_CIRCpedia_342868) was measured to form the strongest circR loop by binding its cognate DNA at the exons 9−10 within the breakpoint cluster region (bcr) of MLL recombinome. Moreover, compared to normal cohort without blood cancers, MLL–rearranged leukemia patients presented significantly increased expression of circMLL(9,10) in their neonatal blood spots. Therefore, circMLL(9,10) was chosen for elaborating the functions of circR loops (Figure 1). Subsequently, based on a series of in vitro and in vivo experiments, the authors confirmed that the overexpression of circMLL(9,10) did not alter the whole transcriptome or alternative splicing of MLL gene, but paused RNA Pol II, promoted DSBs, reorganized chromatin; and inhibited 20S proteasome, ultimately leading to oncogenic chromosomal translocations of MLL and decreased survival rate of HL-60 cell xenograft mice.
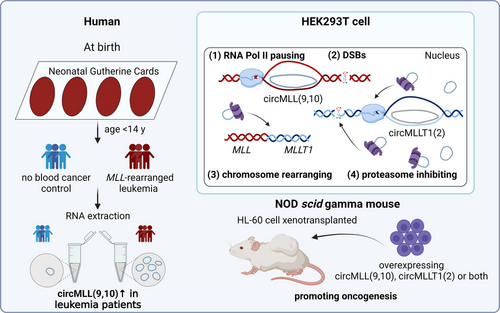
This study by Conn et al. has provided lines of convincible evidence for circRNAs to be genetic risks of tumorigenesis by forming circR loops and impairing genome stability.6 Detecting circRNAs in the neonatal blood spots is a notable highlight, since it was found that circMLL(9,10) significantly increased before the onset of leukemia symptoms, suggesting circRNAs as endogenous RNA carcinogens. Therefore, these circRNAs can be potential candidates as preventive and diagnostic targets. Moreover, the abundance of circRNAs regulates circR loop formation; hence cleaning these circRNAs as a therapeutic approach is conducive to reduce circR loop production and prevent undesired oncogenic gene translocations. A separate study from Chen and colleagues has offered clues to adjust the abundance of nuclear circRNAs. They found that Exportin 4 (XPO 4) was an indispensable circRNA transporter from nucleus to cytoplasm, and deficiency of XPO 4 led to the accumulation of nuclear circRNAs and triggered neurological disorders and fertility defects.10 Based on this finding, we speculate that XPO 4 could be a therapeutic target to eliminate nuclear circRNAs and inhibit circR loop formation. Additionally, the study by Conn et al. indirectly inspires neuroscientists to explore the effects of circR loops in central nerve systems, because the OEA data indicate that circR loops gene set is more significantly enriched in neurological diseases than leukemias.6
ACKNOWLEDGEMENTS
This work is supported by National Natural Science Key Foundation of China (grant numbers: 81830040 and 82130042).
CONFLICT OF INTEREST STATEMENT
The authors declared no potential conflict of interest with respect to the commentary.




