Combinatorial BRD4 and AURKA inhibition is synergistic against preclinical models of Ewing sarcoma
Abstract
Background
Ewing sarcoma (ES), a bone cancer affecting children, adolescents, and young adults, is driven by the EWS-FLI1 fusion protein in the majority of cases, but the mechanisms of tumorigenesis are still being elucidated. Consequentially, therapeutic advances have been limited in the last 25 years. Two therapeutic targets have been recently examined. Aurora kinase A (AURKA) promotes cell cycling and posttranslational stabilization of the oncoprotein MYC, while bromodomain-containing protein 4 (BRD4) promotes gene expression epigenetically. Studies of AURKA and BRD4 inhibitors showed some impairment of tumorigenesis in ES preclinical models, but this efficacy was limited in single-agent use. AURKA and BRD4 activate common oncogenic pathways through different mechanisms, so we hypothesized dual inhibition would be synergistic against tumorigenesis.
Aims
(a) Define the synergistic antineoplastic effects of BRD4 inhibitor I-BET151 and AURKA inhibitor alisertib against ES cell lines in vitro. (b) Confirm the mechanism of activity of each agent in these cell line models. (c) Define the efficacy of I-BET 151 and alisertib alone and in combinations with each other, and with the chemotherapeutic drug vincristine against ES tumor xenografts in vivo.
Methods and results
I-BET151 and alisertib synergistically inhibit viability in ES cell lines SK-ES, TC71, and ES2 in vitro. Alisertib alone upregulates transcriptional expression of its targets, but combined use of I-BET151 mitigates that upregulation, likely contributing to the drug synergy and downregulating RNA and protein expression of key oncogenic pathways as shown by RT-qPCR and western blot. Alisertib and I-BET151 significantly prolong survival in three ES xenograft models in vivo, and this efficacy is augmented by the addition of vincristine.
Conclusion
Dual targeting of oncogenic drivers in ES epigenetically and posttranslationally, specifically by use of BRD4 and AURKA inhibitors, may have significant clinical efficacy in ES alone and/or in combination with current chemotherapy regimens.
Abbreviations
-
- AURKA
-
- aurora kinase A
-
- BRD4
-
- Bromodomain-containing protein 4
-
- CI
-
- combination index
-
- Fa
-
- fraction affected
-
- p-AURKA
-
- phosphorylated AURKA, specifically at threonine 288
1 INTRODUCTION
Ewing sarcoma (ES), a primitive cancer of the bones and soft tissues that affects children, adolescents, and young adults remains a major clinical challenge. Patients with localized disease only at diagnosis have a 30% chance of refractory or relapsed disease,1, 2 and those patients with metastatic disease at diagnosis or relapsed/refractory disease have less than 30% chance of survival.3, 4 A tumor-specific t(11;22) translocation yielding the EWS-FLI1 fusion protein has been identified in 90% of all ES and is the primary oncogenic driver required for viability of these tumors.5-7 EWS-FLI1 had been difficult to target because of its intrinsically disordered structure, though a small molecule that can target it is undergoing evaluation.8 Nonetheless, the mechanism of EWS-FLI1-mediated tumorigenesis is still cryptic, and additional oncogenic pathways must be activated to allow the tumor cells to tolerate EWS-FLI1 expression, as it is toxic to normal tissues.9 Three major genomic studies of ES demonstrated that these tumors harbor 0 to 1 additional mutation at diagnosis,10-12 limiting identification of mutation-activated oncogenic pathways. However, other studies have identified potential oncoproteins that may be therapeutically targetable in ES.
One potential target is aurora kinase A (AURKA), a protein that promotes cytokinesis,13 inhibits p53 function,14 augments passage through the G2/M phase of the cell cycle15, and stabilizes the MYC family of oncogenic transcription factors by preventing FBXW7-mediated ubiquitination.16 AURKA is directly transcriptionally promoted by EWS-FLI1 in ES.17 ES has been specifically shown to be sensitive to therapeutics that target cell cycle progression and cytokinesis,18 suggesting dependence on AURKA expression. This is supported by preclinical work showing knockdown of AURKA by short hairpin RNA decreased ES cell viability.19 The first-in-class drug, alisertib, showed efficacy against ES and select other pediatric cancers preclinically.20 However, in the Phase 1 pediatric clinical trial, alisertib had higher toxicity in children than in adults, limiting its maximally tolerated dose.21 Alisertib failed to meet response criteria in multiple Phase 2 studies when used alone22-25, but next-generation inhibitors of AURKA are in development, and AURKA inhibition in combination with other treatment approaches may be particularly effective against ES.
A second potential target in ES is the bromodomain and extraterminal motif (BET) chromatin-binding protein BRD4. BRD4 recognizes and localizes to acetylated lysine residues,26 promoting transcription through recruitment and phosphorylation of RNA Polymerase II.27 BRD4 is active and oncogenic in cancers by promoting expression of multiple targets, including CDK4/6,28 MCL1,29, 30 BCL2,31 MYC,32, and AURKA.33 BRD4 has been more recently shown to be critical for EWS-FLI1-directed transcriptional reprogramming.34 BRD4 inhibitors showed some preclinical efficacy against ES34-36; while BRD4 inhibition alone could not induce ES tumor regression in these studies, the data did show BRD4 inhibition could inhibit tumor formation.35, 37 These data suggest that BRD4 inhibition may have cytostatic effects that limit its use as a solo agent against ES but may augment other molecular therapeutic approaches.
Inhibition of AURKA and BRD4 affects many common oncogenic pathways active in ES through distinct and complementary mechanisms. In this report, we show that the AURKA inhibitor alisertib and the BRD4 inhibitor I-BET151 have significant synergy against ES cell lines in vitro, inhibiting cell proliferation and viability while using markedly lower concentrations of each drug. We demonstrate that treatment of cells with alisertib causes a “rebound” upregulation of transcription of AURKA and MYC, but combinatorial treatment with I-BET151 mitigates that upregulation. As a result, the drug combination more efficaciously represses protein expression of multiple targets, including AURKA, MYC, CDK4/6, and BCL2, and it also is more effective in repressing AURKA kinase activity. In ES xenograft models, alisertib and I-BET151 more efficaciously extend survival when used together as compared with either drug alone. The inclusion of the antitubulin agent vincristine further augments the efficacy of these drugs. These data suggest a novel therapeutic approach for targeting oncogenic pathways in ES.
2 MATERIALS AND METHODS
2.1 Cell lines
SK-ES, ES2, and TC71 cell lines were obtained from Peter Houghton (Greehey Children's Cancer Research Institute, San Antonio, Texas). All cell lines were authenticated by PowerPlex16 short tandem repeat analysis (Promega) at the start of in vitro studies and again prior to in vivo studies. Cells were cultured in DMEM (Corning, Bedford, Massachusetts) with 10% FBS (PeakSerum, Wellington, Colorado) at 37°C with 5% CO2 and confirmed to be free of Mycoplasma by SouthernBiotech Mycoplasma Detection Kit (Birmingham, Alabama), tested every 3 months.
2.2 Drugs
Alisertib was purchased from ApexBio (Houston, Texas). I-BET151 was obtained from GlaxoSmithKline (Collegeville, Pennsylvania). A list of primers and antibodies used can be found in the supplementary data.
2.3 Cell viability assay, combination index analysis, and LIVE/DEAD assay
SK-ES, ES2, and TC71 cells were plated in 96-well plates at 5000 cells/well, respectively, in complete media in triplicate wells for each dose and cultured for 24 hours. Cells were then treated with either alisertib (dissolved in 100% ethanol) to final concentration in media of 20 to 2000 nM (with 0.1% ethanol total) or vehicle control, or I-BET151 (dissolved in DMSO) to final concentration in media of 200 to 8000 nM (0.1% DMSO total) or vehicle control, to define IC50s for each drug alone at 48 hours of treatment. Cells were then treated with either alisertib (dissolved in 100% ethanol) to final concentration in media of 20 to 1000 nM (with 0.1% ethanol total) and DMSO vehicle, I-BET151 (dissolved in DMSO) to final concentration in media of 200 to 2000 nM (0.1% DMSO total) and ethanol vehicle, both drugs, or vehicle controls for 48 hours. Cell viability was quantified using the IncuCyte ZOOM live cell imaging system (Essen BioScience, Ann Arbor, Michigan) to track percent confluence of each well. Percentage confluence as compared with vehicle control was used to calculate treatment effect. IC50 was calculated using GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, California). Combination index (CI) values were calculated using Compusyn software (Combosyn, Inc., Paramus, New Jersey). Cells were treated at experiment endpoint with the Invitrogen LIVE/DEAD viability/cytotoxicity assay (ThermoFisher Scientific, Waltham, Massachusetts) per manufacturer protocol. In brief, at experimental endpoint, media was removed from the cells and washed with phosphate buffer saline (PBS). The cells were then treated with PBS containing 1 μM calcein AM and 2 μM ethidium homodimer. Viable cells take up calcein AM and dead cells take up ethidium homodimer. Cells were incubated for 45 minutes then imaged using the IncuCyte Zoom with fluorescence imaging settings. Viability was assessed by green fluorescence; cytotoxicity was assessed by red fluorescence. Three independent experiments were performed; representative experiments are shown here.
2.4 Reverse transcription—Quantitative polymerase chain reaction (RT-qPCR)
Cells were grown to 80% confluence and then treated with alisertib (the IC50 concentration for alisertib for each cell line: TC71, 100 nM; ES2, 150 nM, SK-ES, 250 nM,), I-BET151 (the IC50 concentration for I-BET151 for each cell line: TC71, 6000 nM; ES2, 2000 nM; SK-ES, 8000 nM), a combination of the two drugs at those doses, or vehicle control for 24 hours. Total RNA was extracted from the cells using NucleoSpin RNA purification kit (Takara Bio USA, INC, Mountain View, California), and 1 μg of RNA used for cDNA synthesis using Maxima RT cDNA First Strand Synthesis kit (ThermoFisher Scientific). qPCR was performed using KiCqStart SYBR Green qPCR ReadyMix (Sigma-Aldrich, St. Louis, Missouri) using the ABI PRISM 7900HT thermal cycler (ThermoFisher Scientific), with relative quantitation by the ddCt method as previously described.38 Primers used are listed in Table S1. Experiments were performed with technical duplicates on each plate and in three independent experiments, with the relative expression of each experiment used to calculate expression and standard deviation, plotted on each graph.
2.5 Western blot
Cells were grown to 80% confluence, then treated with alisertib (the IC50 concentration for alisertib for each cell line: TC71, 100 nM; ES2, 150 nM, SK-ES, 250 nM,), I-BET151 (the IC50 concentration for I-BET151 for each cell line: TC71, 6000 nM; ES2, 2000 nM; SK-ES, 8000 nM), a combination of the two drugs at those doses, or vehicle control for 48 hours. Cells were collected by rubber policeman and lysed in radioimmunoprecipitation (RIPA) buffer with Halt protease/phosphatase inhibitor complex (ThermoFisher Scientific), with 50 μg of protein/sample used for western blot as previously described.38 Blots were imaged by chemiluminescence using ECL Western Blotting Substrate (ThermoFisher Scientific). Band intensity was quantified using ImageQuant TL software (GE Healthcare, Marlborough, Massachusetts), then normalized by comparing each band with its actin control sample, then to the vehicle control sample to generate a ratio of relative expression. Experiments were performed in independent triplicate; representative images are shown here.
2.6 Tumor xenograft studies
5×106 cells of each indicated type were detached with TrypLE Express solution (ThermoFisher Scientific), resuspended in PBS and then mixed 1:1 in Matrigel (Corning) to a final volume of 100 μL/injection. The suspended cells were subcutaneously injected into the right flanks of SCID mice (Envigo, Indianapolis, Indiana), one injection per mouse. Tumors were allowed to grow to approximately 100 to 200 mm3 as estimated by volume = (length × width2)/2. Mice were then treated with the listed drug combinations, n = 5 per group, with drugs at the following doses and routes: I-BET151, injected intraperitoneally 20 mg/kg/day; alisertib, orally by gavage 20 mg/kg/day; and vincristine, injected intraperitoneally 0.1 mg/kg/dose once weekly (formulations in the Supplementary Methods). We planned to treat mice with drug and/or vehicle for 5 days × 5 weeks, then observe without treatment for 2 weeks (two-drug combination) or 3 weeks (combinations with vincristine); treatment or observation were halted when they reached tumor or humane endpoint as noted below. Mice were evaluated for distress daily by the vivarium staff, independently of the researchers. Mice were also twice-weekly weighed and had tumors measured. Mice were euthanized when tumors attained 2000 mm3, reached humane endpoints, or at study end, with tumors harvested. Mice euthanized for distress have been indicated in the figures. All studies were designed in accordance with Nationwide Children's Hospital Institutional Animal Care and Use Committee (IACUC) guidelines and performed under IACUC-approved protocols.
2.7 Statistical analysis
Statistical analyses were performed using GraphPad Prism 7. Where appropriate, the two-tailed Student's t-test was used to calculate significant differences between comparison groups in the experiments above. For multiple comparisons, one-way analysis of variance (ANOVA) was used with analysis of multiple comparisons of each group to the other. Mantel-Cox log rank test was used for survival analyses. IC50 values and drug synergy in vitro were calculated using Compusyn software, as described above.
3 RESULTS
3.1 Alisertib and I-BET151 synergistically inhibit ES cell growth in vitro
We first evaluated if alisertib and I-BET151 were synergistic against ES in vitro. We used the ES cell lines SK-ES, ES2, and TC71, which all express the canonical EWS-FLI1 fusion protein. We treated the ES cells with alisertib and I-BET151 at a range of concentrations from 0 to 2000 nM for the former and 0 to 8000 nM for the latter for 48 hours, defining individual IC50 doses for cell line alone (Figure S1). All three ES lines demonstrated sensitivity to each drug, though to varying degrees.
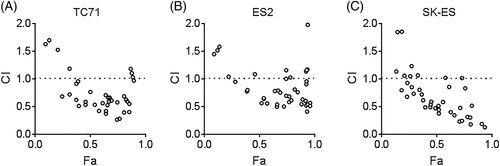
To evaluate for synergistic antineoplastic effects of the drugs, we treated the cells with combinations of the drugs (0-1000 nM for alisertib and 0-2000 nM for I-BET151, as this spanned at least to the IC25 range for each drug) and evaluated cell viability at 48 hours (Figure S2). Using Compusyn software, we found that, in all three cell lines, the drugs demonstrated synergistic effects against proliferation and viability at most drug combinations (Figure 1). Compusyn calculates, for each drug concentration combination, the anticipated effect of each drug alone and compares this with the observed effect, calculating a CI.39 CI < 1 reflects a greater-than-additive combinatorial effect, with CI < 0.7 considered synergistic. For all three ES lines, the combinations with concentrations of I-BET151 > 200 nM and with concentrations of alisertib < 1000 nM demonstrated synergy, with CI < 0.7 across the majority of combinations. These results supported our hypothesis that dual AURKA and BRD4 inhibition would have synergistic antineoplastic effects against ES.
3.2 I-BET151 represses transcriptional expression of genes regulated by AURKA and BRD4 and augments AURKA inhibition in repressing protein expression of these targets
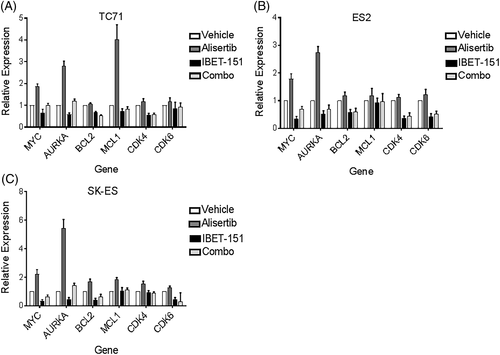
We hypothesized that alisertib and I-BET151 synergize because of their complementary mechanisms of activity. We hypothesized that alisertib, which acts posttranslationally to repress oncoprotein stability and expression, induces a reflexive increase in transcriptional expression of those targets. As such, treatment with I-BET151, which represses transcriptional expression of many of the same targets of alisertib, would be expected to complement alisertib treatment. We evaluated our hypothesis first by examining the transcriptional expression of AURKA, MYC, CDK4, CDK6, BCL2, and MCL1, all of which are transcriptionally driven by BRD4 activity. TC71, SK-ES, and ES2 cells were treated with IC50 doses of alisertib, I-BET151, both, or equal volume of vehicle, for 24 hours, and then RNA expression was evaluated by RT-qPCR (Figure 2). For most of the targets, treatment with I-BET151 repressed gene expression below levels seen in the vehicle-treated cells, as would be expected with BRD4 inhibition, whereas treatment with alisertib alone caused increased expression, particularly of AURKA, MYC, BCL2, and MCL1. Cotreatment of the cells with I-BET151 and alisertib attenuated the compensatory overexpression of these genes (one-way ANOVA of the four treatment groups, for TC71, P = 0.0483; for SK-ES, P = 0.022; for ES2, P = 0.0158, with the alisertib groups significantly different in expression from the other treatment groups for each cell line, P < 0.05).
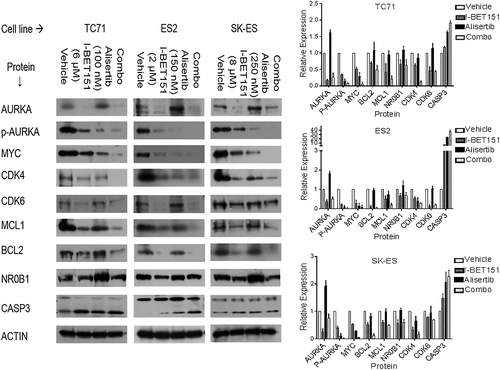
We validated our hypothesis of the drug combination's effects on oncoprotein expression by western blot. We also evaluated the effects of the drug combination on AURKA function, as evaluated by its capacity to autophosphorylate at threonine-288 (“p-AURKA”), on EWS-FLI1-driven protein expression as evaluated by the EWS-FLI1 target NR0B1, and on apoptosis as quantified by CASP3 cleavage (Figure 3). In all three cell lines, we showed that alisertib inhibited AURKA phosphorylation (P < 0.05 all three cell lines), confirming its on-target effects, and this inhibition correlated with decreased MYC expression (P < 0.02). Alisertib treatment also induced CASP3 cleavage in all three cell lines, consistent with the cytotoxic effects seen above (P < 0.01). However, alisertib treatment also induced significantly higher total AURKA expression in all three cell lines, consistent with the RNA expression analysis (P < 0.03). Alisertib had no significant effect on NR0B1 expression in any of the cell lines (P > 0.5). I-BET151 alone, in contrast, reduced total AURKA expression (P < 0.01) and also had correspondingly lower AURKA phosphorylation compared with control, though this reduction in p-AURKA was not as low as alisertib alone (P < 0.02). I-BET151 treatment also repressed protein expression of multiple canonical BRD4 targets, including MYC (P < 0.03 vs control in all three cell lines), CDK4/6 (P < 0.05 vs control in all three cell lines), and more variably BCL2 and/or MCL1. It also significantly repressed NR0B1 expression in all three cell lines (P < 0.05), though to a lesser extent than the other BRD4 targets. Interestingly, I-BET151 treatment did induce CASP3 cleavage in all three cell lines as compared with vehicle control (P < 0.01) but to a lesser degree than alisertib (P < 0.05). The combination of alisertib and I-BET151 prevented upregulation of AURKA as compared with alisertib alone in all three cell lines (P < 0.02), with correspondingly lower p-AURKA in TC71 and SK-ES cells as compared with either drug alone or control (P < 0.05); alisertib alone was as efficacious as the drug combination in reducing p-AURKA in the ES2 cells (P = 0.67). The drug combination was also more efficacious at repressing MYC expression than either drug alone (P < 0.05) with more variable effects on the other BRD4 targets and with no greater repression on NR0B1 than I-BET151 alone. Additionally, the drug combination caused greater CASP3 cleavage than either drug alone in all three cell lines (P < 0.03). These data support our hypothesis that dual AURKA and BRD4 inhibition is efficacious against oncogenic pathway activation in ES.
3.3 Alisertib and I-BET151 are more efficacious against ES tumor xenografts in vivo than either drug alone
These data led us to evaluate the efficacy of the drugs in vivo. TC71, SK-ES, and ES2 cells were implanted as Matrigel plugs subcutaneously in SCID mice, forming tumor xenografts. When the tumors were greater than 100 mm3, the mice were treated with alisertib, I-BET151, both drugs together, or vehicle alone, for 5 weeks or to tumor or humane endpoint, followed by a 2-week drug washout period of observation. All mice tolerated the drug regimens, with no observed stress or significant weight loss. One mouse in the ES2 combination, SK-ES I-BET151, and SK-ES combination treatment groups were euthanized prior to maximum tumor endpoint because of ulcerations in their tumors but were included in the final analysis because these were due to tumor effect.
In the mice with TC71-derived tumors, alisertib treatment extended survival as compared with vehicle (Figure 4A, median survival 26 days vs 20 days, P = 0.027; tumor growth curves Figure S3), as did I-BET151 treatment (median survival 26 days, P = 0.018). I-BET151 and alisertib together were significantly better than either drug alone in extending survival (median survival 31 days, P = 0.040 vs I-BET151, P = 0.042 vs alisertib).
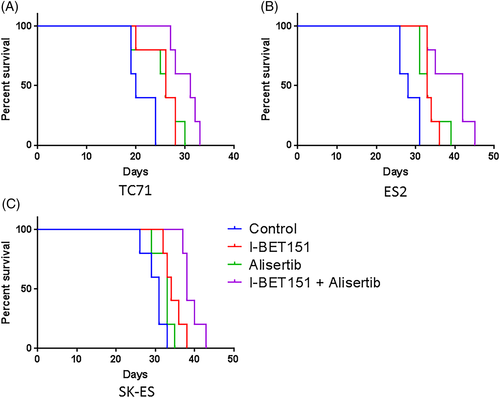
In the mice with ES2-derived tumors, alisertib treatment extended survival as compared with vehicle (Figure 4B, median survival 33 days vs 28 days, P = 0.017; tumor growth curves Figure S4), as did I-BET151 treatment (median survival 33 days, P = 0.0025). I-BET151 and alisertib together were significantly better than either drug alone in extending survival (median survival 42 days, P = 0.046 vs I-BET151, P = 0.038 vs alisertib), with three mice surviving the treatment duration but with tumor progression meeting endpoint during the washout period.
In the mice with SK-ES-derived tumors, alisertib treatment approached significance in extending survival over control (Figure 4C, median survival 33 days vs 31 days, P = 0.08; tumor growth curves Figure S5), whereas I-BET151 treatment did significantly extend survival (median survival 34 days, P = 0.015). I-BET151 and alisertib together were significantly better than either drug alone in extending survival (median survival 38 days, P = 0.022 vs I-BET151, P = 0.0028 vs alisertib), with all five mice treated with both drugs surviving the treatment duration but with tumor progression meeting endpoint during the washout period.
3.4 Vincristine combined with alisertib and I-BET151 is tolerated and further improves antitumor efficacy against ES tumor xenografts in vivo
The results of AURKA and BRD4 inhibition against ES tumor xenografts in vivo supported our hypothesis that this dual inhibition would have improved efficacy against this cancer. However, while the combination of alisertib and I-BET151 promoted apoptosis in vitro, the combination did not induce tumor regression in our models. We further hypothesized that the addition of a rationally-selected chemotherapeutic agent could further improve antitumor efficacy of alisertib and I-BET151. Vincristine, an antitubule vinca alkaloid, has been previously demonstrated to synergize individually with both BRD4 inhibitors40 and AURKA inhibitors41 in other cancers. For this reason, and as further discussed below, we tested the efficacy of vincristine with alisertib, with I-BET151, and with both agents.
We first evaluated the tolerability of this three-drug combination. We found that the mice tolerated vincristine at a dose of 0.2 mg/kg intraperitoneally weekly alone or with alisertib, but there was increased toxicity in mice at that dose in combination with I-BET151, manifested as weight loss and urinary retention. However, the mice tolerated a vincristine dose of 0.1 mg/kg weekly in combination with either alisertib or I-BET151 or both. We then performed an extended xenograft treatment study with 5 weeks of treatment and 3 weeks of drug washout and observation, with treatment or observation ceased when mice reached tumor or humane endpoint. All the mice tolerated this dosing regimen. One mouse in the ES2 three-drug treatment group met humane endpoint prior to tumor size endpoint due to ulcerations in the tumor, but this mouse was included in the final analysis as this early death was due to tumor effect.
In the mice with TC71-derived xenografts, vincristine alone had no significant benefit as compared with vehicle control (median survival 21 days vs 20, P = 0.93, Figure 5A; tumor growth curves Figure S3). The addition of vincristine to alisertib significantly improved survival over vincristine alone (median survival 31 days, P = 0.015) and but did not significantly improve survival as compared with alisertib alone (P = 0.062) or compared with alisertib and I-BET151 (P = 0.3592). Vincristine with I-BET151 did extend survival as compared with vincristine alone (median survival 31 days, P = 0.0071) and as compared with I-BET151 alone (P = 0.0348) but not as compared with alisertib and I-BET151 (P = 0.96). The combination of vincristine, alisertib, and I-BET151 was the most efficacious of all drug combinations (median survival 35 days, P = 0.012 vs vincristine and alisertib, P = 0.039 vs vincristine and I-BET151, P = 0.047 vs alisertib and I-BET151), with two mice surviving into the drug washout period, though with subsequent tumor progression (Figure S3).

In the mice with ES2-derived xenografts, vincristine alone had a modest but significant benefit as compared with vehicle control (median survival 31 days vs 28, P = 0.030, Figure 5B; tumor growth curves Figure S4). The addition of vincristine to alisertib did not significantly improve survival as compared with vincristine alone (median survival 33 days, P = 0.078) or as compared with alisertib alone (P = 0.46). Vincristine with I-BET151 similarly did not significantly improve overall survival as compared with vincristine alone (median survival 34 days, P = 0.15) or as compared with I-BET151 alone (p = 0.46). The combination of vincristine, alisertib, and I-BET151 was the most efficacious of all drug combinations (median survival 48 days, P = 0.0080 vs vincristine and alisertib, P = 0.0018 vs vincristine and I-BET151, P = 0.037 vs alisertib and I-BET151), with all five mice surviving into the washout period and one mouse surviving through the washout period without its tumor reaching endpoint (Figure S4).
In the mice with SK-ES-derived xenografts, vincristine alone had no significant benefit as compared with vehicle control (median survival 32 days vs 31, P = 0.45, Figure 5C; tumor growth curves; Figure S5). The addition of vincristine to alisertib did improve survival over vincristine alone (median survival 36 days, P = 0.032), was not significantly better than alisertib alone (P = 0.11) but was inferior to alisertib and I-BET151 (median survival 38 days, P = 0.025). Vincristine with I-BET151 did extend survival as compared with vincristine alone (median survival 36 days, P = 0.0050) and was not significantly better than I-BET151 alone (P = 0.18) or alisertib and I-BET151 (P = 0.18). The combination of vincristine, alisertib, and I-BET151 was the most efficacious of all drug combinations (median survival 43 days, P = 0.0021 vs vincristine and alisertib, P = 0.0018 vs vincristine and I-BET151, P = 0.033 vs alisertib and I-BET151). Again, all five mice survived the drug treatment period, with two mice surviving through the washout period as well without meeting tumor endpoints (Figure S5). The data from these three mice cohorts all support our hypothesis that the addition of a synergistic chemotherapeutic agent to AURKA and BRD4 inhibitors has efficacy against ES tumors.
4 DISCUSSION
ES is a primitive bone cancer characterized in most cases by expression of the EWS-FLI1 fusion protein.5, 42, 43 While EWS-FLI1 has been acknowledged to the oncogenic driver of ES, it was long considered to be an “undruggable target” because of its structure. Furthermore, the multiple functions of EWS-FLI1 in this cancer are still being elucidated,44-47 prohibiting indirect targeting of just one critical pathway. While candidate agents are in development against EWS-FLI1,8, 48 these alone do not cause tumor regression in preclinical models. Additionally, the paradoxical effects of EWS-FLI1-induced growth arrest,49 apoptosis50, and toxicity51 suggest that additional oncogenic drivers must be activated to allow ES tumorigenesis and progression.52 Those oncogenic drivers could serve as novel therapeutic targets to augment current treatment strategies. Prior studies demonstrated that AURKA17, 19, 20, 53 and BRD434-37, 54 both promote oncogenic phenotypes in ES, making them attractive therapeutic targets. However, preclinical and clinical studies using agents targeting these proteins showed limited efficacy against ES when used alone.21, 36 Our data show that inhibition of AURKA and BRD4 together augment their activity, acting synergistically to repress oncogenic pathway activation and inhibit tumor progression. We confirmed the on-target activity of I-BET151 against BRD4, demonstrating the transcriptional repression of multiple canonical BRD4 targets previously validated in different cancers.31, 55-57 We also confirmed AURKA kinase inhibition by alisertib and concomitant MYC repression, consistent with prior studies.58-60 Our work does not preclude other biological impacts of alisertib and I-BET151 on ES biology; in fact, we do confirm that BRD4 inhibition does impair EWS-FLI1-mediated transcriptional activation as shown by I-BET151's repression of NR0B1, consistent with recent data.34 Nonetheless, our data does demonstrate a mechanism of synergy between the two drugs, further augmented by use of chemotherapy such as vincristine.
While AURKA and BRD4 inhibition had synergistic effects against the ES models, those effects did vary among the ES cell lines used. The TC71 cell line, which has the fastest rates of growth in vitro and as tumor xenografts in vivo, showed the highest sensitivity to alisertib of the three cell lines tested here. The ES2 cell line, which has an intermediate rate of proliferation in vitro and in vivo, showed comparatively lower sensitivity with alisertib and higher sensitivity to I-BET151. The SK-ES cell line, which has the slowest proliferation of the three cell lines tested and slowest tumor growth, was also the least sensitive to each drug. This suggests that hyperproliferative cells may be more sensitive to the inhibition of cytokinesis and cell cycling induced by alisertib. They may also rely more on AURKA to maintain MYC expression to allow cell cycling, as demonstrated by the effects of alisertib on MYC protein expression (Figure 3). In contrast, the slower-growing cells may be more dependent on transcriptional regulation of cell cycling and proliferation, reflecting a greater effect on protein expression by I-BET151 in the SK-ES and ES2 cells. Other mechanisms may also contribute to relative resistance to each drug, including modest effects of either drug on MCL1 in all three cell lines and potential upregulation of other drug resistance mechanisms, such as drug efflux.61 Some of these mechanisms may also be adaptive responses, such as de novo mutations or epigenetic changes, which may explain the variation within treatment groups in the in vivo studies, ie, why some tumors generated from a given cell line and treated with a specific drug combination had stable disease while others generated from the same cell line and treated with the same regimen progressed. Additional studies are warranted as to which survival mechanisms are activated in ES in response to AURKA or BRD4 inhibition specifically and in response to cell stress more generally. This will include further analysis of the tumors from the in vivo studies, including assessment of proliferation and apoptosis, and expanded cohort studies to delineate effects on tumors at different time points. These data will allow for refinement of the drug treatment schedule, improvements in the next generation of inhibitors against these proteins, and/or identification of additional drugs that may augment their effects.
While alisertib demonstrated preclinical activity against pediatric solid tumors including ES, it did not meet study criteria for significant benefit in single-agent use in clinical trials.22, 25, 62 In those studies, because of the myelosuppression caused by the drug at the maximally tolerated dose, it was administered for 7 days followed by 2 weeks of recovery. Our data shows that alisertib treatment causes a transcriptional overexpression of AURKA and MYC, presumptively as a resistance mechanism to the inhibition of AURKA kinase activity. This overexpression may explain the lack of efficacy of alisertib clinically, as cells with increased AURKA and MYC expression may rebound after alisertib washout. Our data also show that BRD4 inhibition can mitigate this rebound effect, which we hypothesize allows for greater antitumor efficacy. This result may impact clinical trial design and identify drug combinations that pair kinase inhibitors, delivered in pulsatile fashion, with transcriptional or epigenetic regulatory drugs, given as steady-state maintenance agents, to impair cell growth and then maintain that repression.
It is noteworthy that, while AURKA protein expression was inhibited in cells treated with alisertib and I-BET151 to lower levels as compared with alisertib alone, it was not as low as I-BET151 alone. This suggests that BRD4-independent mechanisms also regulate and augment AURKA expression, particularly in response to AURKA inhibition; additional work is needed to elucidate those mechanisms.
While alisertib and I-BET151 were significantly more efficacious in vivo together than alone, they did not induce any durable tumor responses, particularly when treatment ended. This may relate to the hyperproliferative nature of established ES cell lines, but it concerned us that there may be limited clinical effect of the drug pair if used in isolation. As combination drug therapy is the cornerstone of treatment of pediatric cancers, we hypothesized that a cytotoxic chemotherapy drug, selected for a mechanism of action that complements AURKA and BRD4 inhibition, would further augment their efficacy. We selected vincristine as a proof of principle. It has been shown previously to be synergistic with BRD4 and AURKA inhibition,40, 41 disrupting cell cycling and sensitizing the cells tested in those studies to apoptosis. Vincristine has demonstrated efficacy in combination therapy for ES clinically and recently been shown to sensitize ES cells to “mitotic catastrophe” through cell cycle disruption.18 However, alisertib and I-BET151 have additional effects on ES biology, and additional studies are needed to evaluate how vincristine-mediated effects interact with BRD4 and AURKA inhibition for antitumor efficacy. These studies will include assessment of microtubule phosphorylation and dynamic, cell cycle arrest, activation of cell death and antiapoptotic pathways, and interplay with EWS-FLI1-regulated biology.
Clinically, vincristine is minimally myelosuppressive, avoiding a known toxicity of alisertib.21 However, we encountered increased toxicity with the combination of I-BET151 and vincristine. While we were encouraged to see efficacy of the three-drug combination using a reduced dose of vincristine, this could be prohibitive clinically. Furthermore, while the three-drug combination did extend overall survival and had some effects on preventing tumor growth, it did not induce tumor regression. Some of these limitations could be due to the methods used, including the relatively small numbers of mice used in each group and the use of ES xenografts from cell lines as opposed to primary patient-derived xenografts (which harbor fewer genomic alterations). Identification of other chemotherapeutics that disrupt cell cycling and/or other BRD4 inhibitors, such as I-BET76263-65 and OTX015,56, 66 next-generation AURKA inhibitors,67 and/or other drugs in these classes, may lead to a more tolerable and efficacious combination therapy. Additional studies on these tumors, including transcriptomic and proteomic evaluations of the tumors in response to drug treatment and evaluation of clinical biomarkers of disease severity such as Ki-67 expression, would further elucidate the mechanisms of activity of these drugs in isolation and in combination. Finally, as prior studies with BRD4 inhibitors showed efficacy in preventing tumor growth, future in vivo studies will include evaluation of prevention of metastasis formation and disease control in an adjuvant setting, which may have more relevant clinical applications in frontline disease treatment.
This study shows that therapies that target common oncogenic pathways both transcriptionally and posttranslationally can be synergistic. Posttranslational inhibition may induce rapid antiproliferative changes and apoptosis, but these effects may be limited in their duration. Targeting of gene expression, either through transcriptional disruption such as with BRD4 inhibition or epigenetic modification by histone deacetylase or methytransferase inhibitors, may induce a more durable inhibitory effect on cancer cells but can be slow to have effect on cell proliferation and viability. Dual targeting of gene expression and protein function and stability may improve clinical efficacy and may salvage drugs that have failed primary clinical trial endpoints in single-agent use. These drugs can be dose-adjusted to allow better tolerability while increasing efficacy. These approaches can be specifically adapted for use in ES, to augment current combination chemotherapy and radiation therapy strategies, to maintain disease control in the setting of metastatic or unresectable disease, or to augment therapies targeting EWS-FLI1 activity.
5 CONCLUSIONS
Our data show that the AURKA inhibitor alisertib and the BRD4 inhibitor I-BET151 complement each other in ES to inhibit cell proliferation and viability, promote apoptosis, and to extend survival in preclinical models of disease. The mechanisms of activity of this combination therapy, through transcriptional and posttranslational inhibition of oncogenic pathways, are more efficacious together than independently, and this is further augmented by the antitubule chemotherapeutic vincristine, which additionally disrupts cell cycling and cytokinesis. This work supports further study of dual BRD4 and AURKA inhibition in ES, both in combination therapy with additional chemotherapeutics and with novel agents, including those targeting EWS-FLI1.
ACKNOWLEDGMENTS
We thank Olena Barbash at GlaxoSmithKline for her assistance and expertise in the use of the BRD4 inhibitor I-BET151 in vitro and in vivo.
CONFLICTS OF INTEREST
The authors declare on conflict of interest.
AUTHOR CONTRIBUTIONS
All authors had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Conceptualization, N.S.; Methodology, L.T., E.B., J.F., J.S.A., N.S.; Investigation, L.T., E.B., J.F., J.S.A., N.S.; Formal Analysis, L.T., E.B., N.S.; Resources, N.S.; Writing—Original Draft, L.T., E.B., N.S.; Writing—Review & Editing, N.S.; Visualization, N.S.; Supervision, N.S.; Funding Acquisition, N.S.
FUNDING INFORMATION
This work was supported by the Families for a Cure foundation (grant number 20064014) and Cancer Free Kids (grant number 82178416).




