Individualized tacrolimus therapy: Insights from CYP3A5 polymorphisms and intestinal metabolism
Key Clinical Message
CYP3A4 and CYP3A5 are the most abundant and important enzymes of the CYP3A subfamily, distributed in the liver, intestinal mucosa and kidney, and involved in tacrolimus metabolism. Here, we report a case of tacrolimus dosage refractoriness due to a genetic polymorphism of CYP3A5.
1 INTRODUCTION
The Pharmacokinetics of tacrolimus are different in each case and require an individualized administration design based on blood concentration measurement (therapeutic drug monitoring: TDM). Main attention has been focused on the importance of drug metabolism in the liver or extraction from the kidney, but absorption in the intestinal tract is also a major factor. Genetic polymorphisms exist in CYP3A5, which is involved in the metabolism of tacrolimus, including *3 and *1 alleles. The combination of these alleles affects the blood concentration of tacrolimus. The *1 allele carriers have high metabolic activity, and the *3 allele carriers have low metabolic activity. These metabolisms occur not only in hepatocytes but also in intestinal epithelial cells. We showed here two characteristic cases in which was difficult to control the blood concentration of tacrolimus due to the CYP3A5 gene polymorphism, probably in the intestinal tract.
2 CASE HISTORY
Case 1: A 50-year-old female visited our hospital for a living donor kidney transplant. In 2012, a medical examination revealed hypertension and renal dysfunction with a serum Cr level of 3.1 mg/dL, and she was referred to our hospital. The right kidney was hypoplastic, and the left kidney was atrophic and the kidney was uneven surface at CT images. With clinical manifestation of limited amount of proteinuria and CT images, the case was clinically diagnosed as hypoplastic kidney and nephrosclerosis.
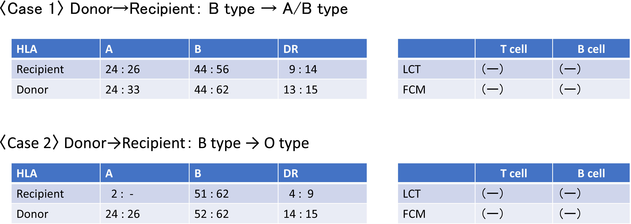
Her renal function deteriorated over time, and her serum Cr level worsened to 8.0 mg/dL in July 2015, she was placed on hemodialysis in February of the same year. In December 2017, she was scheduled to undergo an ABO-compatible living donor kidney transplant (Table 1: recipient: A/B type, donor: B type, HLA 4 mismatch) with her husband as the donor. Postoperative biopsies showed no acute rejection.
Physical examination revealed that her height was 158 cm, body weight was 67.4 kg, and BMI was 26.9. Her blood pressure was 108/62 mmHg, and her pulse rate was 92 beats/min at administration. Laboratory data showed that Red blood cells were 3.65 million/μL, Hb 11.5 g/dL, and WBC 5970/μL. PLT 240,000/μL, Na 141 mmol/L, K 5.6 mmol/L, corrected Ca 9.5 mg/dL, BUN 58 mg/dL, Cr 10.1 mg/dL, Alb 3.9 g/dL, LDH 155 U/L, AST 9 U/L, ALT 5 U/L.
A standard immunosuppression therapy (methylprednisolone, basiliximab, tacrolimus, and MMF) was started 4 days before transplantation. Tacrolimus was started at a dose of 5 mg/day, but her blood tacrolimus concentration remained lower than 3 ng/mL. Thus, a genetic polymorphism test for CYP3A5 by long-range PCR based targeted next generation sequencing1 revealed that she was an intensive metabolizer with a homozygous *1 allele.
Case 2: A 50-year-old male visited our hospital for a living donor kidney transplant. He was detected with hematuria and proteinuria around 10 years ago. He was treated for chronic glomerulonephritis by a home doctor. Kidney function deteriorated gradually, and serum Cr level reached 10.9 mg/dL. About 10 months after starting hemodialysis, he was referred to our hospital for an ABO-incompatible living donor kidney transplant from his wife (Table 1: recipient: O type, donor: B type, HLA 5 mismatch). Kidney biopsy was not possible at the time of referral, because the kidney function was advanced and the kidneys were atrophic. Postoperative biopsy showed no acute rejection. Physical examination revealed that his height was 175.8 cm, and his weight was 71.5 kg, and BMI was 23.1. His blood pressure was 134/70 mmHg, and his pulse rate was 81 beats/min at administration. Laboratory data showed that Red blood cells were 3.61 million/μL, Hb 11.3 g/dL, and WBC 3470/μL. PLT 161,000/μL, Na 141 mmol/L, K 4.1 mmol/L, corrected Ca 9.6 mg/dL, BUN 31 mg/dL, Cr 7.54 mg/dL, Alb 3.8 g/dL, LDH 139 U/L, AST 8 U/L, ALT 7 U/L. Anti-B antibody: IgM 16-fold, IgG 32-fold.
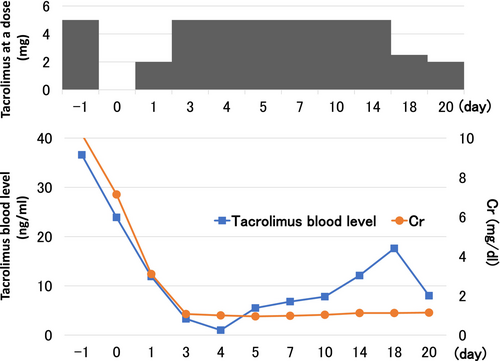
In addition to a standard immunosuppression therapy (methylprednisolone, basiliximab, tacrolimus, and MMF), rituximab was started 14 days before transplantation. Serum antibody titers were less than 32-fold, so plasma exchange was performed once. Tacrolimus was started at a dose of 10 mg/day before transplantation, and our target trough blood concentration was 10 ng/mL during the perioperative period. However, on the second day of oral administration, the trough blood concentration of tacrolimus was markedly high (36.3 ng/mL) (Figure 1). To investigate the cause of the high concentration of tacrolimus, CYP3A5 gene polymorphisms were evaluated, and he had an enzyme deficiency (homozygous *3 alleles).
3 METHODS
Tacrolimus blood levels were measured 48 h after initiation of dosing or dose changes.
The half-life of tacrolimus was calculated using the following formula; t1/2 = ln (2) × (t2−t1)/ln(C1/C2).
Genetic analysis was performed by NGS analysis using the vLAS (very long amplicon sequencing) method: the CYP3A5 genomic region was amplified, and this PCR product was fragmented and fully sequenced by MiSeq. The bases at the relevant SNP loci were then confirmed and genotyped.1
3.1 Differential diagnosis
The drug concentration is associated with drug interactions and intestinal absorption. Since tacrolimus is primarily metabolized by the drug metabolizing enzymes CYP3A4 and CYP3A5, concomitant use with other drugs that are metabolized by CYP3A4 and CYP3A5 may increase blood concentrations of tacrolimus. In addition, concomitant use with drugs that induce CYP3A4 and CYP3A5 may decrease the blood concentration of tacrolimus. Neither of two cases took drugs that affect tacrolimus metabolism. In addition, diarrhea may increase the blood concentration of tacrolimus by allowing tacrolimus to rapidly pass through the upper part of the small intestine, where CYP expression is high, and escape metabolism, but there was no persistent diarrhea in CASE 2, so the effect of diarrhea is unlikely.
3.2 Outcome and follow-up
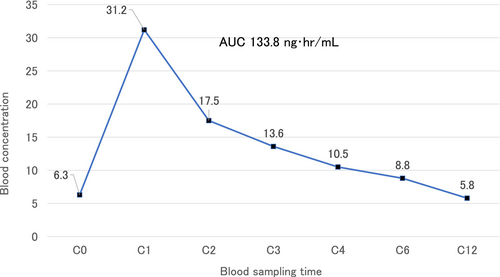
Case 1: A standard immunosuppression therapy (methylprednisolone, basiliximab, tacrolimus, and MMF) was started 4 days before transplantation. Tacrolimus was started at a dose of 5 mg/day, but her blood tacrolimus concentration remained lower than 3 ng/mL. Thus, the dose of tacrolimus was increased to 12 mg/day, the trough tacrolimus concentration levels reached the optimal value (around 10 ng/mL). The half-life of tacrolimus in blood was 10 h, which was within the standard range (Figure 2).On the 10th day of transplantation, the AUC of tacrolimus at a dose of 12 mg oral administration was 133.8 ng・h/mL (Figure 3).
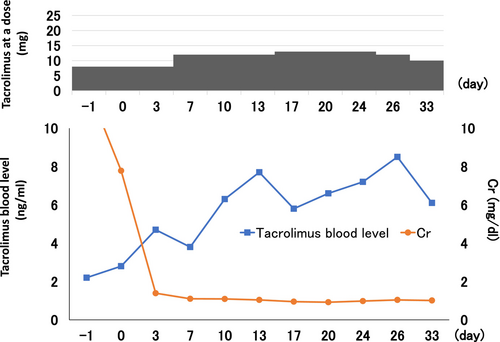
Case 2: Tacrolimus was started at 10 mg/day before transplantation, with a target perioperative trough blood level of 10 ng/mL. However, on the second day of oral administration, the trough blood concentration of tacrolimus became significantly higher (36.3 ng/mL), and tacrolimus was discontinued for 2 days. The blood half-life of tacrolimus was 22.1 h, which was expected due to delayed metabolism and excretion of tacrolimus; after 2 days of discontinuation, the dose of tacrolimus was reduced to 5 mg and the target trough blood concentration (5–8 ng/mL) was achieved. Subsequently, blood levels were easily elevated, and trough blood levels were measured and adjusted accordingly.
4 DISCUSSION
Here, we report two cases in which a particularly high dose of tacrolimus and a particularly low dose of tacrolimus were required to control the optimal blood concentration of tacrolimus. In both two cases, the half-life of the blood concentration was within the standard range. Therefore, there were no major abnormalities in the metabolism in the liver. On the other hand, genetic polymorphism testing for CYP3A5 revealed that one patient was homozygous for the *1 allele, which indicates an intensive metabolizer, and the other was *3 allele (Table 2), which indicates an enzyme-deficient type.
| NG_007938.2 (ATG start) | 3699C>T | 6981A>G | 12,947T>C | 14660A>G | 14685G>A | 19381G>A | 27126_27127insT | 27284C>A |
|---|---|---|---|---|---|---|---|---|
| Case 1 | ||||||||
| Effect on protein (NP_000768.1) | p.R28C | Splicing defect | Splicing defect | p. Q200R | Splicing defect | p. A337T | p. T346fs | p. T398N |
| Position at NC_000007.14 (GRCh38.p13) | g. 99676198G>A | g. 99,672,916 T>C | g. 99666950A>G | g. 99,665,237 T>C | g. 99665212C>T | g. 99660516C>T | g. 99652771dup | g. 99652613G>T |
| dbSNP_ID | rs55817950 | rs776746 | rs55965422 | rs56411402 | rs10264272 | rs28383479 | rs41303343 | rs28365083 |
| CYP3A5 Allele *1 (Wild type) | G | T | A | T | C | C | A | G |
| CYP3A5 Allele *2 | T | |||||||
| CYP3A5 Allele *3 | C | R (A or G) | Y (C or T) | Y (C or T) | K (G or T) | |||
| CYP3A5 Allele *4 | C | |||||||
| CYP3A5 Allele *5 | G | |||||||
| CYP3A5 Allele *6 | T | |||||||
| CYP3A5 Allele *7 | AA | |||||||
| CYP3A5 Allele *8 | A | |||||||
| CYP3A5 Allele *9 | T | |||||||
| Sample Allele *1/*1 | G/G | T/T | A/A | T/T | C/C | C/C | A/A | G/G |
| Case 2 | ||||||||
| Effect on protein (NP_000768.1) | p.R28C | Splicing defect | Splicing defect | p.Q200R | Splicing defect | p.A337T | p. T346fs | p.T398N |
| Position at NC_000007.14 (GRCh38.p13) | g.99676198G>A | g.99672916T>С | g.99666950A>G | g.99665237T>C | g.99665212C>T | g.99660516C>T | g.99652771dup | g.99652613G>T |
| dbSNP ID | rs55817950 | rs776746 | rs55965422 | rs56411402 | rs10264272 | rs28383479 | rs41303343 | rs28365083 |
| CYP3A5 Allele *1 (Wild type) | G | T | A | T | C | C | A | G |
| CYP3A5 Allele *2 | T | |||||||
| CYP3A5 Allele *3 | C | R (A or G) | Y (C or T) | Y (C or T) | K (G or T) | |||
| CYP3A5 Allele *4 | C | |||||||
| CYP3A5 Allele *5 | G | |||||||
| CYP3A5 Allele *6 | T | |||||||
| CYP3A5 Allele *7 | AA | |||||||
| CYP3A5 Allele *8 | A | |||||||
| CYP3A5 Allele *9 | T | |||||||
| Sample Allele *3/*3 | G/G | C/C | A/A | T/T | C/C | C/C | A/A | G/G |
According to genetic polymorphism testing for CYP3A5, these two cases were found to be homozygous for the *1 allele, which indicates an intensive metabolizer type, and the *3 allele, which indicates an enzyme-deficient type. Tacrolimus is absorbed in the gastrointestinal tract, reaches the liver via the portal vein, and is metabolized primarily by CYP3A4 and CYP3A5.2 CYP3A4 present in the liver degrades various medicines to metabolites such as the 13-O-demethylated form, which is excreted into bile.3 On the other hand, CYP3A5, which has a similar substrate specificity to CYP3A4, also contributes to the degradation of tacrolimus.4 CYP3A5 has genetic polymorphisms, that are *1 and *3. CYP3A5 *3/*3 cases is a deficiency in the CYP3A5 enzyme protein (enzyme-deficient type).5, 6 In Japan, it is reported that 7.0% of patients are intensive metabolizers (*1/*1) and 60.5% are poor metabolizers (*3/*3).7 The CYP3A5 *3/*3 patient in Case 2 does not have CYP3A5 activity and tacrolimus is mainly metabolized by CYP3A4.5 In addition, it has been reported that in cases of extensive metabolizers, as in Case 1, the dose of tacrolimus is 1.5–2 times higher than in normal cases.8 Therefore, the reason for the significant difference in dosage of tacrolimus in the two cases would be the difference in the genetic background of CYP3A5 (homozygotes for the *1 and *3 alleles).
In these two cases, the doses of tacrolimus were significantly different, but the half-life in the blood, which reflects the metabolism in the liver, was standard range (Case 1; 10 h, Case 2; 25.5 h, standard range, 8–40 h). In drug metabolism, in addition to the liver, which is involved in drug metabolism in the blood, intestinal metabolism, which is involved in absorption, is also important. Drugs absorbed from transporters in intestinal epithelial cells depredate within the epithelial cells. The amount of CYP in the small intestine is not uniform.9, 10 Total CYP content increases slightly from the duodenum to the jejunum and then decreases toward the ileum.11 Extensive metabolizers such as CYP3A5*1/*1 degrade tacrolimus in intestinal epithelial cells, making it difficult to absorb tacrolimus into the bloodstream. In other words, due to metabolism by intestinal CYPs, orally administered drugs are largely eliminated before reaching systemic circulation. In these cases, the blood half-life of tacrolimus was within the standard range. Therefore, it was speculated that the difference in the required dose of tacrolimus was due to absorption in the gastrointestinal tract rather than liver metabolism. Based on the above discussion, it is speculated that the main cause of the significant difference in the doses of tacrolimus in these cases is the metabolism of tacrolimus by CYP3A5 in the gastrointestinal tract (intestinal epithelial cells).
In conclusion, we believe that it is useful to evaluate genetic polymorphisms in CYP3A5 in patients before kidney transplantation to make a drug administration plan for tacrolimus.
AUTHOR CONTRIBUTIONS
Mizuki Mishima: Writing – original draft. Tomohisa Yabe: Conceptualization. Takaya Kondo: Conceptualization. Keiji Fujimoto: Conceptualization; writing – review and editing. Ryoji Takata: Conceptualization; data curation. Hitoshi Yokoyama: Conceptualization. Yo Niida: Conceptualization; formal analysis; writing – review and editing. Tatsuro Tanaka: Conceptualization; writing – review and editing. Katsuhito Miyazawa: Conceptualization; writing – review and editing. Kengo Furuichi: Conceptualization; resources; writing – review and editing.
FUNDING INFORMATION
This research received no spe`cific grant from any funding agency in the public, commercial, or not-for-profit sectors.
CONFLICT OF INTEREST STATEMENT
The authors state that the study was conducted without any commercial or financial relationships that could be interpreted as a conflict of interest.
ETHICS STATEMENT
All procedures performed in studies involving human participants were by the ethical standards of the institutional committee and with the 1964 Helsinki Declaration and its later amendments or comparable thical standards.
CONSENT
Written informed consent was obtained from the patient to publish this report in accordance with the journal's patient consent policy.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no data were created or analyzed in this study.