Expression assay of the COLQ in a family with congenital myasthenic syndrome and symptomatic carriers
Mohammad Farid Mohammadi and Sahand Tehrani Fateh have contributed equally to this work.
Key Clinical Message
Congenital myasthenic syndromes-5 (CMS5) is a rare autosomal recessive heterogeneous disorder, caused by pathogenic variants in the COLQ that lead to skeletal muscle weakness and abnormal fatigability. The onset is usually from birth to childhood. Disease-causing variants in the collagen-like tail subunit are the most explained etiology in synaptic CMS, causing defected acetylcholinesterase. In this study whole-exome sequencing (WES) was performed in an affected boy with muscle weakness, ophthalmoplegia, and bilateral ptosis and gene expression assay by qRT-PCR was performed in entire family. A homozygous nonsense variant in the COLQ [NM_005677.4:c.679C>T], (p.Arg227Ter) was identified in the proband. Segregation analysis by Sanger sequencing confirmed the homozygous state in the proband and heterozygous state in his parents and four of the siblings. The mRNA expression level in the proband was 0.02 of a healthy person, and in the carriers were 0.42 of a healthy person. This study presents an Iranian family with two affected children and eight symptomatic carriers with attenuated mRNA expression. This study provides evidence that carriers of the COLQ disease-causing variants could become symptomatic with some yet unknown pathogenesis mechanism and underscore the importance of further investigations to elucidate this mechanism.
1 INTRODUCTION
Congenital myasthenic syndromes (CMSs) are a heterogeneous group of disorders characterized by defects affecting neuromuscular junctions (NMJ). CMSs are classified based on their genetic and clinical characteristics into presynaptic, synaptic basal-lamina, and postsynaptic.1 CMSs as a group of genetic disorders with mostly autosomal recessive inheritance, usually present in infancy or childhood. Family history of CMS and absence of acetylcholine receptor (AChR) antibodies, distinct CMS from autoimmune myasthenia gravis (MG).2, 3 Despite the low incidence of CMSs, a genetic diagnosis should be considered seriously in cases with myasthenic syndrome. The exact distinction of CMS from MG is of great importance in treatment, as CMS cases usually do not respond to anti-acetylcholinesterase (AChE) therapy, plasma exchange, or thymectomy. Additionally, medications that might be effective for one type of CMSs can be harmful in another type. While AChE therapy, such as pyridostigmine and amifampridine, are beneficial for some types of CMS, they clinically worsen the cases of AChE deficiency. Treatment with ephedrine and albuterol could be effective in some cases of CMS5.4, 5 Considering all together, genetic testing is essential for distinguishing CMS from MG and also differentiating different types of CMS in order to choose the appropriate treatment for each patient.
So far, total of 35 protein-encoding genes with essential functions at the neuromuscular junction (NMJ) have been identified to cause CMS.2, 6-9 Pathogenic variants in the CHRNE are responsible for more than half of the cases in the United States.10 Moreover, variants in RAPSN, CHAT, COLQ, and DOK7 have been reported in many cases.6 Muscle contractions are stimulated by the release of acetylcholine (ACh) and terminated by AChE which degrades the ACh.11 AChE is an enzyme concentrated at the endplate of the neuromuscular junction and anchors into the basal lamina by a collagenic tail. The collagenous tail of AChE12, 13 is encoded by the COLQ located on 3p25.1, and disease-causing variations in this gene cause CMS5 (OMIM: 603034). CMS5 is characterized by severe and progressive muscle weakness, often beginning at birth or early childhood; however, late-onset cases manifest a milder course of the disease. In CMS5, associated reported symptoms are ophthalmoparesis, ptosis, poor feeding, and respiratory insufficiency due to muscle weakness.8, 14, 15 However, it is worth pointing out that the features mentioned above are only consistent in some patients. Although the mode of inheritance is autosomal recessive in this disease, some carriers of mutations in the COLQ have been reported with CMS5 manifestations but milder symptoms.16
Due to the importance of differentiating different types of CMS from each other and MG, genetic testing is a necessity for patients with related clinical findings. Additionally, phenotypic variation in severity and symptoms could be found in CMS5 patients, and even milder symptoms have been reported in the carriers of COLQ variants. This paper reports a COLQ variant found by WES in an Iranian patient with synaptic CMS and investigates COLQ expression by qRT-PCR in the affected patient and eight symptomatic relatives who were carriers of the same variant.
2 METHODS
2.1 DNA isolation
Whole-blood genomic DNA was extracted from the affected proband and his healthy family members using the standard protocol. Nanodrop and agarose gel electrophoresis were used to measure the concentration and quality of the extracted DNA.
2.2 Whole-exome sequencing and bioinformatics analysis
Extracted DNA was sequenced by WES using Illumina HiSeq 4000 platform (average coverage depth > 100X). Reads were aligned to (GRCh38/hg38) using BWA software.17 We used ExAc, Iranome, dbSNP, GnomAD, and 1000Genome project databases to check variants with minor allele frequency <1%. An in-house bioinformatics pipeline was used for bioinformatic analysis. Prioritization and filtering of the relevant variants were performed manually based on similar studies.18-20 Variants were excluded by manual filtering on the annotated VCF file, considering the inheritance mode, clinical information, and phenotypes according to HPO terms (https://hpo.jax.org/app/), consequence and allele frequency of variants according to 1000Genome project, GnomAD, and ExAC databases. After that, we classified the remaining variants according to the American College of Medical Genetics and Genomics (ACMG) guideline rules.21
2.3 Mutation validation and segregation analysis
Sanger sequencing and segregation analysis were performed to confirm the variant found in the proband and all other family members, using Applied Biosystems 3130 Genetic Analyzer. Primers were designed by the PrimerQuest tool (https://www.idtdna.com/) and the blat tools of the UCSC genome browser (https://genome.ucsc.edu/).22 Sequence results were analyzed by Codoncode aligner software to detect the variants.23
2.4 RNA isolation
Ten milliliters of blood were collected from the patient and the entire family members in sodium EDTA-containing tubes. Total RNA isolation was performed on whole blood using the RNeasy Midi Kit, Qiagen. Concentration (range of 0.6–2.3 μg/μl) and purity was measured via Nanodrop (BioTech) (OD260/280 ratio = 1.8–2).
2.5 Reverse transcription
We used 1 μg of extracted RNA for reverse transcription via QuantiTect Reverse Transcription Kit, Qiagen. The integrity and quality of template cDNA were evaluated by cDNA amplification of GAPDH as an internal control reference gene.24 Primer Quest tool was used to design qRT-PCR primers (Table 1).
| Gene | Forward primer 5′–>3 | Reverse primer 5′–>3 | Amplicon length (bp) | Accession number | Tm |
|---|---|---|---|---|---|
| COLQ | GGAGGAGTGTGACGACGGTA | TCTCGCATGTCAGGTAGCCA | 137 | NM_080539.4 | 61–62 |
| GAPDH | ATGGAGAGTAGTACAACAGCCTC | CATGAGTCCTTCCACGATACC | 116 | NM_001357943.2 | 61–62 |
2.6 QRT-PCR assay
According to the high expression of this gene in peripheral blood,25 we examined the expression of the COLQ mRNA by qRT-PCR. QuantiTect® SYBR® Green PCR Kit, Qiagen in CFX96 qRT-PCR detection system (BioRad) was used to assay the COLQ gene expression in the proband and all his family members. We used negative and positive controls for assay quality control of these steps. Each reaction was performed in the final volume of 50 μL: 25 μL 2x Master Mix, 2 μL template cDNA, and 10 picomoles of each forward and reverse primer. The first phase of the qRT-PCR program was initial denaturation (95°C for 5 min) and then forty cycles of denaturation (94°C for 15 s); the second phase was annealing (55°C for 30 s), and the third phase was extension (72°C for 30 s). We analyzed melting to check the accuracy of the COLQ and GAPDH amplification considering the melting temperature.
2.7 Statistical analysis
All statistical analyses were conducted by the SPSS software (V26).26 We used the Student's t-test and ΔΔCt method to check gene expression changes. In this study, we considered p-value <0.05 as significant.
3 RESULTS
3.1 Case presentation
A 15-year-old boy born to consanguineous parents, was presented with muscle weakness with onset symptoms in infancy. He was unable to run and jump during the first years of his life. His parents noticed him having difficulty smiling, speaking, chewing, and swallowing for a long time, in addition to droopy eyelids. As he grew up, he required assistance to do his physical activities, such as climbing stairs. Early diagnosis of “myasthenia gravis” was made at the age of 10. The Tensilon test at the time was negative. Pyridostigmine was prescribed for him, but his weakness got worse. Hence, this medication was discontinued. Fortunately, he adapted well, and remained autonomous in his daily activities. His further clinical evaluations revealed bilateral ptosis and ophthalmoparesis in all gazes. Slow pupillary response to light was not found. The gag reflex was fine. Muscle forces of proximal and distal of both upper and lower extremities were three out of five. He had axial muscle weakness; however, with no signs of scoliosis or kyphosis. Deep tendon reflexes were within the normal range. Mild unsteadiness was detected in his gait. No sensory deficits or sphincter dysfunction were detected. No thymus enlargement was found. Biochemical tests were normal, including fasting blood sugar (FBS), CBC, renal, liver, thyroid function, and other markers, the results of the anti-AChR and anti-muscle-specific tyrosine kinase (MuSK) antibodies were negative. Sensory nerve conduction, motor conduction velocity, and electromyography (EMG) represented myasthenic syndrome, and decremental response following repetitive nerve stimulation (RNS) was observed. Double compound muscle action potential (CMAP) in response to single nerve stimulus was evident. By investigating the family history, it was revealed that he had an older brother who had severe symptoms of myasthenia, such as hypotonia, difficulty in sucking and swallowing, and neurodevelopmental delay, who died at the age of 2 years. Clinical examinations revealed mild ptosis and swallowing abnormality in proband's father and mother. Additionally, mild symptoms, including mild ptosis and muscle weakness, were found in some other family members, as shown in Figure 1A.
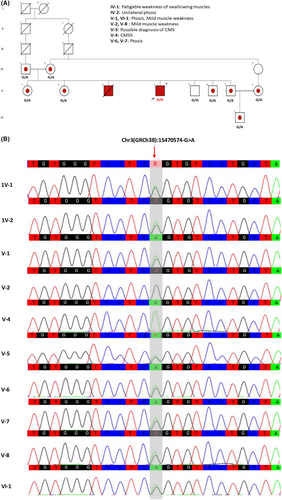
3.2 Exome sequencing analysis and Sanger sequencing validation
WES was performed on the 15-year-old affected proband and after bioinformatic analysis, we found a homozygous pathogenic variant in the COLQ: [Chr3(GRCh38):15470574-G>A, NM_005677.4:c.679C>T, (p.Arg227Ter). This variant has been reported in homozygous state in two other patients and in compound heterozygous state (with p.Val332Asp and p.Arg410Trp) in two other patients.8, 15, 27, 28 Segregation analysis by Sanger sequencing confirmed the homozygous state in the proband and heterozygous state in their parents and four of the siblings. The pedigree and the Sanger chromatogram of the variant are shown in (Figure 1A,B).
3.3 Expression levels of GAPDH as a reference gene
Ct value of GAPDH was calculated in samples to assay the reference gene expression in the whole family. The GAPDH Ct values in the family members were between 21.17 and 22.50.
3.4 Expression levels of COLQ
We evaluated ΔCt of GAPDH and COLQ through the following method: [Ct value of reference gene – Ct value of COLQ] to assay the expression levels of COLQ. For the healthy member, ΔCt was (−5.73), the mean of ΔCt for carrier members was (−6.97), and ΔCt of the proband was (−11.37). Then, for evaluating ΔCt changes between healthy, carrier, and proband, we calculated ΔΔCt of COLQ by use of the following method: ([ΔCt of proband – ΔCt of healthy] and [mean ΔCt of the carrier- ΔCt of healthy]). The results were (−5.64) for proband, (−1.24) for the carrier, and 0 for healthy. According to the Livak method, we calculated the E−ΔΔCt value to assay the expression fold change level of the patient, healthy, and carrier members.29 The expression level in the proband was 0.02 of a healthy person, and in the carriers were 0.42 of a healthy person (Figure 2).

4 DISCUSSION
Congenital myasthenic syndrome-5 (CMS5), also referred to as end-plate acetylcholinesterase deficiency (EAD), is caused by homozygous or compound heterozygous pathogenic variants in the COLQ. This study reports the [NM_005677.4: c.679C>T; (p.Arg227Ter)] variant in an affected boy. This variant leads to a premature stop codon and is classified as pathogenic based on the recommendations of ACMG. The COLQ spans approximately 50 kb and comprises 17 constitutive exons and a couple of alternatively transcribed exons that encode a collagen-like strand that associates with two other COLQ protein to form a triple helix to make a tail that anchors catalytic subunits of AChE to the basal lamina. The COLQ gene is expressed in 124 organs, and the highest expression levels are within the uterine tube, heart ventricle, skeletal muscle tissues, and whole blood. The COLQ protein has 455 amino acids and contains two collagen-like domains, including, collagen-like 1 (position 96–269) and collagen-like 2 (position 277–291), the proline-rich attachment domain (PRAD) binds the AChE catalytic subunits (position 51–67), two heparan sulfate proteoglycan binding domain (HBS1: position 130–133, HBS2: position 235–238), trimerization domain (Trm) and cysteine-rich domain. The c.679C > T, (p. Arg227Ter) variant either causes nonsense-mediated decay (NMD) in transcripts or leads to the formation of truncated proteins. According to the qPCR result, the mRNA level of COLQ protein in the affected patient is just 0.02 of the control; hence, the NMD of transcripts is more probable than the truncated protein's expression. However, if transcripts escape NMD, the outcome of the c.679C>T, (p. Arg227Ter) variant is the truncation of 227–455 residues which eliminates the major domains of the protein which were responsible for the formation of the triple helix, including collagen-like, PRAD, HBS, Trm, and c-terminal of the protein. Hence, even if transcripts escape NMD, this variant dramatically disrupts the 3D structure and performance of the protein, and as a result, the T-isoform of AChE cannot be linked to the triple-helical COLQ tail.30
According to the type of the variant, COLQ pathogenic variants can manifest from the neonatal period to late childhood. In this patient, clinical symptoms were severe and started from infancy, probably due to the lack of vital domains of the protein. COLQ protein interacts with NMJ and skeletal muscle proteins, including PRIMA1, BCHE, PLOD2, ACHE, CHRNE, DOK7, MUSK, RAPSN, CHRNA1, and HSPG2, which have an essential role in signal transduction and the normal function of skeletal muscles, suggesting the significant impact of COLQ in these pathways.
Patients with pathogenic COLQ variants usually present with muscle weakness and fatigability with onset between infancy and adulthood, which increases with exertion.31 Poor feeding and respiratory difficulties could be present due to weakness of muscles involved in swallowing and respiration. Ocular muscle involvement is common among patients with CMS5, and it usually presents as bilateral ptosis; however, asymmetric ptosis has also been reported in some cases.8 Hypotonia, dysarthria, scoliosis and lordosis, ophthalmoparesis, and delayed pupillary light reflex have also been reported as clinical findings of CMS5 patients; however, it is worth mentioning that features mentioned above are not consistent in all patients, and phenotypic variation in severity and symptoms could be seen.15, 32, 33 The main clinical symptoms in our patient were delayed motor milestone, bilateral ptosis, ophthalmoparesis, generalized weakness due to a defect at the neuromuscular junction exacerbated by exertion, and difficulty in smiling, speaking, and chewing for an extended time. Delayed motor milestone, ptosis, facial weakness and proximal weakness are the shared symptoms between our patient and two other patients that had been reported with the (p.Arg227Ter) variant in the COLQ (Table 2).
| Case | Mutation | Sex | Ethnic origin | Age of onset | Delayed motor milestones | Respiratory crises | Ptosis/ophthalmoparesis/facial weakness/dysphagia | Slow pupillary light response | Proximal weakness/waddling gait/scapular winging/atrophies | Distal muscle weakness/selective involvement of wrist and finger extensors | Axial/neck muscles weakness/ scoliosis or kyphosis | Abnormal tendon reflexes | Progressive disease course | Disease severity | RNS decrement | Double CMAP | Myopathic potentials | Negative or ambiguous response to acetylcholinesterase inhibitors | PMID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Arg227Ter/Arg227Ter | F | Pakistanian | <1 y | + | − | +/−/+/− | N.D. | +/−/−/− | −/− | +/+ | − | − | Mild | − | N.D. | + | ± | 18180250 |
| 2 | Arg227Ter/Arg227Ter | M | Kurdish | 2 y | − | − | +/+/+/+ | − | +/−/−/N.D. | +/N.D. | +/− | N.D. | − | Mild | + | + | + | + | 22088788 |
| 3 | Arg227Ter/Val322Asp | M | N.D. | at birth | + | + | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | + | + | N.D. | + | 23553736 |
| 4 | Arg227Ter/Arg410Trp | M | Caucasian and Filipino descent | 6 m | + | −/−/−/− | N.D. | +/N.D./N.D./N.D. | N.D. | +/+ | N.D. | + | Severe | + | N.D. | N.D. | + | 25557462 | |
| This study | Arg227Ter/Arg227Ter | M | Iranian | <1 y | + | − | +/+/+/+ | − | +/+/−/− | +/N.D. | +/− | − | − | Mild | + | + | + | + |
- Note: ± denotes initial positive response followed by ineffectiveness and worsening.
- Abbreviations: CMAP, compound muscle action potential; F, female; M, male; m, month; N.D., no data; RNS, repetitive nerve stimulation; y, year.
Interestingly, eight individuals of proband's family, including his mother and father, who were carriers for (p.Arg227Ter) variant, showed mild symptoms, including ptosis and muscle weakness. Given that almost all carriers in this family showed some symptoms of CMS, we surveyed the COLQ mRNA expression level in all the family members. As we expected, the expression level in symptomatic heterozygotes was about half (0.42) of the healthy person, but the expression level was deficient in the affected patient (0.02 of the healthy person). Deficient expression in the patient is probably due to the destruction of mRNA by nonsense-mediated mRNA decay (NMD), an mRNA quality control mechanism that degrades mRNAs that harbor a premature termination codon.34
Although CMS5 cases have been reported in populations with different ethnicities, reports of symptoms in carriers of the COLQ variants are very scarce. In a study conducted in 2007, a proband was presented with homozygous c.950delC variant, which leads to a frameshift in exon 13 and truncates the collagen-encoding region of the COLQ protein. The proband had congenital ptosis, muscular weakness, and difficulties in pulmonary adaption at birth. Later in early infancy, delayed gross motor development, muscular weakness, myopathic facies, and external ophthalmoplegia became apparent. Proband's father and paternal grandmother, who were carriers for (c.950delC) variant, also suffered from congenital ptosis.16 Another symptomatic carrier (c.375delT) was reported by Ohno et al. with repetitive compound muscle action potential.35 The underlying mechanism by which carriers become symptomatic is still unclear. Gene dosage effect could explain this phenomenon, taking the carrier symptoms is milder than affected patients; however, many asymptomatic carriers with attenuated mRNA expression have been reported. The dominant negative effect is another mechanism that could explain this phenomenon in which a truncated or modified protein disrupts the activity of normal COLQ protein. A third hypothesis is the presence of a modifying factor that affect the outcome of the COLQ variants.16 Further functional studies are needed to elucidate the underlying mechanism of COLQ variant pathogenesis in the carriers of the COLQ variant.
In conclusion, our study presents an Iranian family with two affected children and eight symptomatic carriers with attenuated mRNA expression. This study provides evidence that carriers of the COLQ variants could become symptomatic with some yet unknown pathogenesis mechanism and underscore the importance of further investigations to elucidate this mechanism. It also emphasizes the importance of comprehended clinical examination and genetic testing in close relatives of probands with COLQ variant as some mild presentations might get missed in COLQ carriers.
AUTHOR CONTRIBUTIONS
Mohammad Farid Mohammadi: Conceptualization; data curation; formal analysis; investigation; methodology; writing – original draft. Sahand Tehrani Fateh: Investigation; methodology; writing – original draft. Hadi Aghajani: Formal analysis; investigation; methodology. Afshin Bahramy: Conceptualization; data curation; formal analysis. Seyed Mohammad Salar Zaheryani: Conceptualization; data curation. Javad Behroozi: Data curation; formal analysis. Seyyed Mohammad Kahani: Data curation; investigation. Pouria Mohammadi: Conceptualization; data curation; formal analysis; methodology. Masoud Garshasbi: Conceptualization; data curation; formal analysis.
ACKNOWLEDGMENTS
We thank the patient and his family for their assistance and contribution. The authors are especially thankful to the personnel of the DeNA laboratory (https://dna-lab.ir/) for supporting us in this study and thank the staff of the Medical Genetics Department of Medical Sciences Faculty, Tarbiat Modares University, Tehran, Iran.
FUNDING INFORMATION
No funding.
CONFLICT OF INTEREST STATEMENT
There is no conflict of interest.
ETHICS STATEMENT
Ethics approval for the study protocol was confirmed by The Human Ethics Committee of Tarbiat Modares University. Written consent was obtained from parents as legal guardians of the proband. The study was performed following the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
CONSENT
Participants' families agreed on the anonymous publication of patients' clinical information and their relevant data.
Open Research
DATA AVAILABILITY STATEMENT
The data are provided upon request.




