Mosaic partial pericentromeric trisomy 8 and maternal uniparental disomy in a male patient with autism spectrum disorder
Key Clinical Message
Various chromosomal anomalies including small supernumerary marker chromosome (sSMC) and Uniparental disomy (UPD) have been described in association with intellectual disability and autism spectrum disorder. Based on our reported findings, we recommend that patients with sSMC(8) be evaluated for autism spectrum disorder (ASD) for early institution of therapy. In the presence of an identifiable sSMC, exploration of UPD is also recommended to further investigate the role of chromosome 8 UPD in ASD.
Introduction
This is a case report of a male patient affected by autism spectrum disorder (ASD) with an 11 Mb mosaic trisomy of the pericentromeric region of chromosome 8 and maternal uniparental disomy (hUPD/iUPD) of the same chromosome. Uniparental disomy (UPD) is defined by the presence or inheritance of a pair of homologous chromosomes from one parent in a diploid genome 1. UPD is classified as (i) heterodisomic (hUPD), denoted by the presence of both uniparental homologues, (ii) isodisomic (iUPD), denoted by the presence of two copies of one homologue, or (iii) a mixture (hUPD/iUPD) of both 2. A rescued meiosis nondisjunction error associated with meiotic recombination and possible additional somatic exchange between chromatids can result in the mixed (hUPD/iUPD) forms 3.
Autism spectrum disorder is a lifelong neurodevelopmental disorder characterized by deficits in social communication and interaction, repetitive and restrictive behavior, extensive clinical and etiologic heterogeneity, as well as, a remarkably rising global prevalence rate. A number of studies have provided evidence supporting the particular role of UPD in ASD, epilepsy and intellectual disability (ID) 4. Furthermore, these neurodevelopmental conditions are also commonly associated with de novo constitutional copy number variations (CNVs) and/or complex chromosomal rearrangements affecting single or multiple chromosomes 5.
Clinical Picture
The proband is currently six-year-old born to nonconsanguineous parents of Greek descent with a birthweight of 3.8 kg. The pregnancy was complicated by polyhydramnios, and dilatation of the pelvicalyceal system identified by ultrasound and the latter resolved spontaneously at birth. He had a normal growth pattern but had gastroesophageal (GE) reflux for the first few months of life. There was difficulty in breastfeeding during the first 2 months but was exclusively breastfed till the age of 15 months. At 18 months, he developed an allergy to cow milk (IgE = 334 IU/mL). He developed chronic asthma with virally induced exacerbations but grew out of it at 4 years of age. His hearing was tested by brain stem evoked potential at the age of 2 years due to delayed speech and was found to be normal. He has hypermetropia in the left eye, and patching the right eye improved visual acuity. He talked at 3 years with dysarthria probably secondary to the hypotonia. Speech therapy for the last 2 years resulted in significant improvement in comprehensible speech. However, he uses short sentences with word repetition and mimicking. An evaluation at the age of 3 years showed that he cannot use a tricycle and can throw a ball but cannot kick it. He could feed himself but was not toilet trained yet. He is impulsive and gets easily frustrated. Another evaluation at the age of 5 years showed that he could follow the orders of three words, and his fine motor and gross motor skills were at 4 years. He could copy 3D objects in 3D, has no concept of money, and does not dress himself but became toilet trained. He has generalized hypotonia and significant drooling secondary to excessive salivation. His electrocardiogram (ECG) and echocardiogram were reported as normal. He has hyperphagia, and at the age of 6 years, his weight is 28 kg (75th %ile), height is 123 cm (90th %ile), and the head circumference is 55 cm (more than two SD above the mean for age and gender).
The proband had two assessments for his neurodevelopment and was diagnosed with mild mental retardation without designation of an IQ figure, attention deficit hyperactivity disorder (ADHD), and pervasive developmental disorder-not otherwise specified (PDD-NOS), those based on Griffith scale. The diagnosis of PDD-NOS is due to lack of communication and abstract thinking, temper tantrums, poor interaction with other children, and obsession with buttons, as well as based on the Autism Diagnostic Observation Schedule (ADOS).
Cytogenetic Evaluation
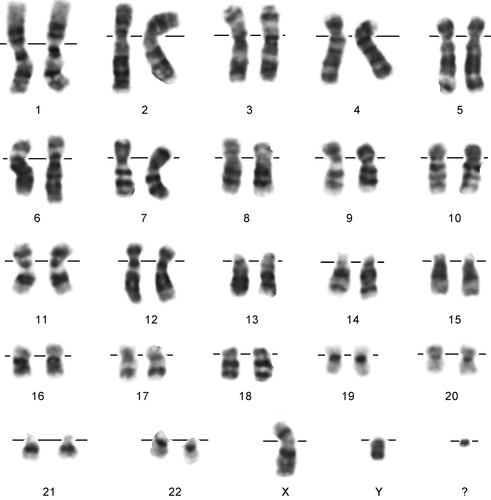
G-banded chromosomal analysis showed a mosaic (60%) marker chromosome (47,XY,+mar) (Fig. 1). Parents have normal chromosomal complement. Cytogenetic analysis method is described in the Supporting Information.
Molecular Evaluation
DNA was extracted from peripheral blood of the proband and his parents after obtaining an informed consent. The consents and the research project were approved by the Qatar Biomedical Research Institute Institutional Review Board (QBRI-IRB), which operates according to the accords of the declaration of Helsinki. Fragile-X syndrome was ruled out by molecular analysis.
Genomewide SNP genotyping was performed, utilizing the OmniExpress array (700K) on an Illumina (San Diego, California) platform. Genotyping quality assessment, SNP calling, and copy number variation (CNV) analysis were performed using the GenomeStudio V.2011 software (human genome build 37/Hg19). The analysis showed the absence of paternal contribution to the child's chromosome 8, except for an 11.29 Mb duplicated region, thus indicating both a partial pericentromeric trisomy of chromosome 8 and a maternal uniparental disomy. The pericentromeric trisomy (38,989,813–50,283,147; p11.22-q11.21) is present in the proband but not in the parents [de novo]. The region is demarcated by the markers (rs4733980, rs7001086) and contains 38 protein-coding genes. A complex pattern of mixed heterodisomy and isodisomy (hUPD/iUPD) indicated by the intermittent regions of loss of heterozygosity (LOH) (172,939–17,483,118 and 77,032,017–119,348,323) and heterozygosity (17,483,118–38,989,813 and 50,283,147–77,032,017) on the child's chromosome 8 was also observed. This chromosomal pattern changed with further molecular evaluation using microsatellite genotyping analysis.
To validate the identified trisomy, quantitative real-time PCR (qRT-PCR) was performed using the SYBR® Green PCR Master Mix on a 7900 fast real-time PCR system (Applied Biosystems, Foster City, California). Primer sets were designed to amplify two exons in genes (ADAM18, exon 12, 393 bp and EFCAB1, exon 2, 403 bp) and three intronic regions (chr8:28.2 Mbp, 172 bp; 38.98 Mbp, 402 bp; 50.28 Mbp, 391 bp) located on the p and q arm, using the web-based tool Primer3 (http://bioinfo.ut.ee/primer3/) (Table S1). The exonic DNA fragments within the trisomic region showed two copies in the parents and three copies in the child. The three intronic DNA fragments peripheral to the trisomy showed two copies in the parents and the child, thus confirming the trisomy (Fig. 2).
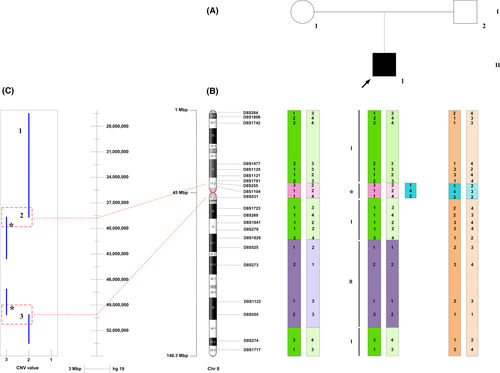
Microsatellite marker analysis was performed using 48 polymorphic markers spanning chromosome 8 to determine the parental origin of the identified regions of complex disomy (hUPD/iUPD). The genotypes of the child and both parents for 21 informative microsatellite markers (Table S2) showed homozygosity in the child but not in the mother in one LOH region (75,958,125–137,687,554), thus confirming maternal isodisomy for this region. Conversely, identical heterozygous fragment sizes in the mother and affected child confirmed the observation of maternal heterodisomy at the rest of the chromosome including region (172,939–17,483,118), which was suggested by the SNP array to be isodisomic. Maternal and paternal inheritance was noted for markers within the pericentromeric region of trisomy (Fig. 2).
The identified CNV was systematically compared to CNVs present in the Database of Genomic Variants (DGV) (http://dgv.tcag.ca/dgv/app/home) to assess its frequency in control populations. The CNV was considered novel if it did not coincide with any CNVs reported in the database. No duplications matching or overlapping with the trisomy identified here were found in control population studies upon searching the DGV database. Furthermore, the identified CNV was compared to CNVs reported to cause known chromosomal imbalance syndromes that are documented in the DECIPHER GRCh37 database (https://decipher.sanger.ac.uk/index). Multiple reports of overlapping duplications on chromosome 8 in patients with comparable phenotypes, including seizures, autistic features, and intellectual disability were identified (Table 1).
| CNV | Inheritance | Size (Mb) | Clinical phenotype | Cases | Chromosomal makeup |
|---|---|---|---|---|---|
| Gain chr8:35115442-51502226 | Unknown | 16.39 | Cleft palate, seizures, small for gestational age | 1 | Unknown |
| Gain chr8:42347178-53861129 | De Novo | 11.51 | Abnormal immunoglobulin level, feeding difficulties in infancy, intellectual disability, microcephaly, proportionate short stature, recurrent infections, short attention span | 1 | 46,XY |
| Gain chr8:39035948-62777195 | De novo | 23.74 | Abnormality of the stomach, febrile seizures, intellectual disability, motor delay | 2 | 46,XX |
| Gain chr8:39245798-39405724 | Unknown | 0.16 | Clinodactyly of the 5th finger, delayed speech and language development, fine hair, high anterior hairline, microcephaly, prominent ears, tapered finger | Unknown | 46,XX |
| Gain chr8:42492810-4706215 | Unknown | 4.57 | Intellectual disability, seizures | 1 | 46,XY |
| Gain chr8:38303653-48872505 | De novo | 10.57 | Autism, severe global developmental delay | 1 | Unknown |
| Gain chr8:37228320-43364903 | Unknown | 6.14 | Intellectual disability | 1 | 46,XX |
| Gain chr8:38764909-39884619 | Unknown | 1.12 | Absence seizures, autism, intellectual disability, delayed speech and language development, frontal upsweep of hair | 1 | 46,XY |
| Gain chr8:42303650-42447498 | Unknown | 0.144 | Global developmental delay, seizures | 2 | 46,XY |
- The clinical phenotype, number of cases, and chromosomal makeup for each reported duplication are shown.
Using the SFARI database for ASD candidate genes 6, a number of chromosome 8 genes that have been implicated in various forms of de novo or inherited ASD and/or intellectual disability were identified (Table S3).
Discussion and Conclusion
Simultaneously occurring, multiple chromosomal abnormalities are usually rare in a single patient 2. Mixed (hUPD/iUPD) is most often observed in unidentified small supernumerary marker chromosome (sSMC) cases 7. For a significant number of UPD cases, undetected mosaicism, recessive genetic disease, and pathogenic structural rearrangement have been identified 8. The collective effect of these events possibly explains the observed clinical phenotypes in this patient, which directly depends on the gene content of the chromosome involved. For the majority of chromosomes, UPD is without phenotypic effects. However, in certain chromosomal segments, it can result in clinically recognizable phenotypes due to imprinting effects the result of inherited differences in gene expression 9. A role for this mechanism is not suspected in this case, as evidence supporting the presence of imprinted genes on chromosome 8 has not been reported 10. Another disease-causing mechanism in UPD is reduction to homozygosity of a disease variant in the iUPD region, for which the parent of origin is heterozygous. Evidence of this mechanism was noted in a male patient with the autosomal recessive chromosomal instability disorder, Nijmegen breakage syndrome (NBS), due to maternal isodisomy of chromosome 8 11. There are four genes (FABP5, SDC2, VPS13B, and EXT1) within the iUPD region, which have been previously implicated in autism 12-15, but we did not explore this mechanism in this report.
Based on literature evidence, the role of sSMCs in ASD is well documented and should be examined here. ASD, developmental delay and mental retardation were noted, respectively, in 22, 83, and 39% of the 18 sSMC(8) cases without UPD(8) reported in the literature 16. Although 70% of sSMC carriers are clinically asymptomatic, duplications extending past the pericentromere to 8p11.22 or to 8q11.22 have been particularly identified in association with developmental delay and ASD 17, 18. Table S4, summarizes the nine pericentromeric sSMC(8) cases reported specifically with ASD among other neurologic and physical presentations. Two of these cases were found to display small supernumerary ring chromosome 8 19, 20. Six of the nine cases displayed varying degrees of cognitive impairment and developmental delay with no physical malformations. However, cases 3, 4, and 9 were reported with both cognitive delay and physical findings including polydactyly (case 3), dysmorphic facial features (case 4), overgrowth syndrome, scoliosis, single palmar crease, umbilical hernia, and deafness (case 9) 19, 21. Only case 7 was found to display the same phenotypic features noted in our case including developmental delay, ASD, and hypotonia 22. Case 7 describes a 6-year-old male with a de novo, mosaic, chromosome 8-derived sSMC (38.94–47.85 Mb; p11.23→q11.1). The pericentromeric derived sSMC shares 8.9 Mb of the 11 Mb trisomic region described in our patient, which explains the similarity in phenotypic presentation among the two cases.
Several genes contained within the trisomic region of our patient have been previously linked to neuronal function. In particular, CHRNB3 (cholinergic receptor, nicotinic, beta 3), which encodes the subunits of neuronal cholinergic receptors, has been reported within trisomies identified in several patients with rare, de novo ASD-specific CNVs 23.
Approximately, 1.3% of sSMC cases present with UPD 24, 25. There are several reported cases of SMC(8) coincident with complex, maternally, or paternally derived (hUPD or iUPD) 7. To our knowledge, none have been described in association with ASD; however, various degrees of developmental delay, intellectual disability, and dysmorphism were reported in these cases, summarized in Table S5, 11, 26-28. Since to date no known phenotypes are associated with UPD(8), it is plausible that UPD(8) does not contribute to the clinical phenotype in our patient.
In this present case, the observation of three haplotypes at the centromere is suggestive of a trisomy rescue from a meiosis-I nondisjunction, therefore resulting in maternal hUPD 24. The region of isodisomy possibly reflects a meiotic, postzygotic recombination event; however, further investigation is required to validate these speculations. Furthermore, UPD resulting from trisomy rescue is often associated with placental or fetal mosaicism. However, it is challenging to isolate the specific clinical effects of the UPD from those caused by the mosaic trisomy.
As such, this is the first report of a patient with ASD and ID presenting with a complex chromosomal, maternal UPD and sSMC(8). Despite reports of their contribution to the observed phenotype, the exact mechanisms underlying these chromosomal aberrations are not entirely clear. Based on our reported case and on review of literature findings, we recommend testing for sSMC(8) in patients presenting with ASD, developmental delay, and hypotonia. In the presence of an identifiable sSMC(8), exploration of UPD is also recommended to achieve better understanding of phenotype–genotype correlation pattern.
Acknowledgments
This study was funded by Qatar National Research Fund (QNRF) NPRP 6-359-3-095, as well as the Shafallah Foundation Center. We extend our sincerest gratitude to the family investigated for their cooperation. Written informed consent was obtained from the guardian of the patient for publication of their individual details. The consent form is held by the first author's institution and is available for review by the Editor-in-Chief.
Conflict of Interest
The authors would like to declare that they have no competing interests.




