LINC00908 Inactivates Wnt/β-Catenin Signaling Pathway to Inhibit Prostate Cancer Cell Stemness via Upregulating GSK3B and FBXW2
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 81672551, 81872089, and 82102799), Natural Science Foundation of Jiangsu Province (No. BK20210230), Doctor of Entrepreneurship and Innovation in Jiangsu Province (No. JSSCBS20210138), The General Project of Medical Research of Jiangsu Health and Wellness Committee (No. M2020049), 2020 Provincial Natural Youth Project (No. 2008085QH358), the subject was Molecular mechanism of TAM exosomes derived miRNAs regulating EMT in prostate cancer cells; First Affiliated Hospital of Bengbu Medical College Science Fund for Outstanding Young Scholars (No. 2019BYYFYYQ09); Changzhou Health Commission Project (ZD2020034); “512 Talent Cultivation Program” of Bengbu Medical College (by51202306).
Han Guan, Qiang Hu, and Lilin Wan contributed equally to this study.
ABSTRACT
Background
Chemotherapy and androgen-deprivation treatment are the curative approaches utilized to suppress prostate cancer (PCa) progression. However, drug resistance and metastasis are extensive and hard to overcome even though remarkable progress has been made in recent decades. The cancer stem cell-related theoretical model explains the distinct molecular characteristics of cancer, its relapse, metastasis, and drug resistance. Meanwhile, noncoding RNA functions in the formation of drug resistance and metastasis in most cancers. The long intergenic nonprotein coding RNA 908 (LINC00908) has been reported to restrain cell proliferation, migration, and invasion of some cancers like triple-negative breast cancer, diffuse large B-cell lymphoma, PCa, and so on. However, its role in stemness for PCa remains unclear.
Methods
We delved into the impact of LINC00908 in PCa cell stemness and the principal molecular mechanism. Then, the impact of LINC00908 on PCa cell stemness and its corresponding mechanism was explored by using functional assays and bioinformatics evaluation.
Results
We found that LINC00908 was low-expressed in PCa cells, and it exerted suppressive functions in PCa cell stemness and tumor growth. Additionally, we revealed that LINC00908 down-regulation was mediated by the HDAC2-p300-YY1 transcription complex. Moreover, LINC00908 up-regulated glycogen synthase kinase 3 beta (GSK3B) via sponging miR-3179. Meanwhile, LINC00908 deployed DEAD-box helicase 3 X-linked (DDX3X) to facilitate the stabilization of F-box and WD repeat domain containing 2 (FBXW2) mRNA. Importantly, LINC00908 enhanced GSK3B and FBXW2 expression to induce the ubiquitination of β-catenin protein, leading to Wnt pathway inactivation.
Conclusion
These results reveal that LINC00908 inhibits PCa cell stemness via inactivating the GSK3B/FBXW2-regulated Wnt pathway, which might enrich people's knowledge of PCa stemness and provide some new potential biomarkers for PCa.
Abbreviations
-
- 3′UTR
-
- 3′untranslated regions
-
- ActD
-
- actinomycin D
-
- ceRNA
-
- competing endogenous RNA
-
- ChIP
-
- chromatin immunoprecipitation
-
- CHX
-
- cycloheximide
-
- CSCs
-
- cancer stem cells
-
- DDX3X
-
- DEAD-box helicase 3 X-linked
-
- FBS
-
- fetal bovine serum
-
- FBXW2
-
- F-box and WD repeat domain containing 2
-
- FISH
-
- fluorescent in situ hybridization
-
- GAPDH
-
- glyceraldehyde-3-phosphate dehydrogenase
-
- GSK3B
-
- glycogen synthase kinase 3 beta
-
- HDAC2
-
- histone deacetylase 2
-
- HEK293T
-
- human embryonic kidney 293 T
-
- LINC00908
-
- long intergenic nonprotein coding RNA 908
-
- lncRNAs
-
- long noncoding RNAs
-
- miRNAs
-
- microRNAs
-
- Mut
-
- mutant type
-
- NC
-
- negative control
-
- PCa
-
- prostate cancer
-
- PRAD
-
- prostate adenocarcinoma
-
- RIP
-
- RNA binding protein immunoprecipitation
-
- RT-qPCR
-
- quantitative reverse transcription polymerase chain reaction
-
- SD
-
- standard deviation
-
- WT
-
- wild type
1 Introduction
Prostate cancer (PCa) ranks as the second most prevalent cancer that happens in the reproductive and urinary systems among men in the world [1, 2]. The pathogenesis of PCa involves multiple factors, such as age, race, familial heredity as well as androgen levels [3]. Surgery, radiation, hormonal ablation, and chemotherapy are routine therapies for PCa [4]. However, failures often occur after these clinical treatments due to the unclear molecular pathogenesis of PCa, especially in drug resistance and metastasis [5]. Cancer stem cells (CSCs) are a reservoir of cancerous cells that present properties of auto-renewal and are responsible for the initiation and progression of tumors, especially the functions of drug resistance and metastasis of cancer cells [6]. CD133 and CD44 are two well-characterized biomarkers for CSCs [7, 8]. Given that prostate CSCs are of great importance for PCa progression [9], we plan to elucidate the molecular mechanism of action underlying the stemness of PCa cells.
Long noncoding RNAs (lncRNAs) are a type of nonprotein coding transcripts exceeding 200 nucleotides in length [10]. Nevertheless, growing evidence has demonstrated that lncRNAs exert important functions in controlling chromatin remodeling, transcriptional as well as post-transcriptional regulation [11]. Moreover, abnormally expressed lncRNAs take part in cancer cell proliferation, migration, invasion, apoptosis, drug resistance, and so on [12]. Recently, many lncRNAs have been identified to be important regulators in PCa [13]. LncRNA PVT1 plays an oncogenic role and predicts poor prognosis in PCa [14]. LncRNA MEG3 impedes PCa progression via the miR-9-5p/QKI-5 axis [15]. Long intergenic nonprotein coding RNA 908 (LINC00908) has been reported to be low-expressed in human lung adenocarcinoma [16] and triple-negative breast cancer [17]. In addition, LINC00908 may serve as potential prognosis biomarkers for glioma [18]. More importantly, Fan et al. [19] have found that LINC00908 repressed cell proliferation, migration, and invasion in PCa. Nevertheless, the correlation of LINC00908 with PCa cell stemness remains obscure.
The Wnt pathway is a key molecular cascade responsible for regulating multiple cellular processes such as proliferation, differentiation, polarity, and cell stemness, and has also been tightly associated with cancer [20]. The β-catenin is an important effector that triggers the transcription of Wnt-target genes in the nucleus to affect tumor cells [21]. When the Wnt pathway is inactivated, cytoplasmic β-catenin can be degraded [22]. Increasing evidence has suggested that lncRNAs participate in PCa progression through modulating the Wnt/β-catenin pathway [23]. Hence, the relationship between LINC00908 and the Wnt pathway in PCa should be further elucidated, and we explored the association of LINC00908 with PCa cell stemness.
Here, we targeted investigating the function of LINC00908 in PCa cell stemness. LINC00908 was low-expressed in PCa cells and exerted suppressive functions in PCa cell stemness and tumor growth. Then, we found that LINC00908 enhanced GSK3B and FBXW2 expression to induce the ubiquitination of β-catenin protein, resulting in Wnt pathway inactivation. This study reveals that LINC00908 inhibits PCa cell stemness by inactivating the GSK3B/FBXW2-regulated Wnt pathway, which provides new potential biomarkers for PCa.
2 Results
2.1 LINC00908 Suppresses PCa Cell Stemness
Based on GSE70769 data, 155 long noncoding RNAs were found to be linked to relapse-free survival in patients with prostate adenocarcinoma (PRAD) (Table S1). Meanwhile, HR (hazard ratio) values for 65 lncRNAs were smaller than 1 (p < 0.05). In these lncRNAs, we found LINC00908 had a specific high relation to PCa cell stemness based on TCGA data (Figure 1A). Furthermore, LINC00908 inhibits cell proliferation, migration, and invasion of PCa [19], but its role on PCa cell stemness remains to be further explored. CSCs (cancer stem cells) possess the capacities of self-renewal and pluripotent differentiation. Thus, CSCs play crucial roles in tumor carcinogenesis, metastasis, and drug resistance [24]. To disclose the function of LINC00908 in PCa cell stemness, we initially investigated the expression profile of LINC00908 in PCa. Based on TCGA and GEPIA database (http://gepia.cancer-pku.cn/), LINC00908 was low expressed in PRAD samples compared to normal control (Figure S1A–C). Then, cancer stemness was computed by RNAss. LINC00908 not only has a significant negative relation to PRAD stemness, but also plays a critical role in prognosis for PRAD (HR = 0.73, p < 0.05) (Figure 1A,B, Figure S1A).
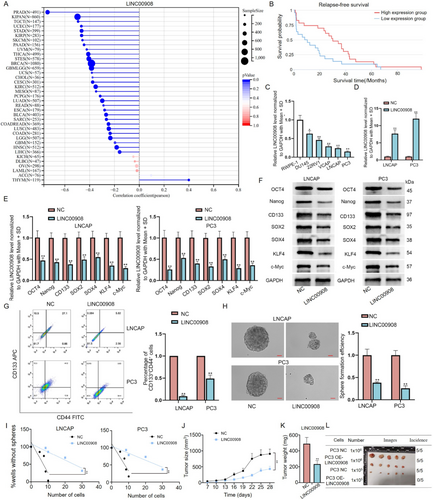
According to RT-qPCR analysis, we discovered that LINC00908 level was substantially lower in PCa cells (DU145, 22RV1, VCAP, LNCAP, and PC3) relative to human normal prostatic epithelial cells (RWPE-1), with a more prominent decline in LNCAP and PC3 cells (Figure 1C). Before gain-of-function assays, we affirmed that the expression of LINC00908 in LNCAP cells increased with the dose of LINC00908-overexpression plasmids (Figure S1D). On this basis, we then transfected 2 μg of LINC00908-overexpression vector into both LNCAP and PC3 cells to realize LINC00908 up-regulation (Figure 1D). Next, through RT-qPCR and Western blotting analyses, we ascertained that increased LINC00908 expression suppressed the levels of several stemness-linked factors (Oct4, Nanog, CD133, SOX2, SOX4, KLF4, and c-Myc) in LNCAP and PC3 cells (Figure 1E,F). Flow cytometry analysis presented the percentage of CD44+CD133+ cells reduced after LINC00908 overexpression (Figure 1G). Moreover, results from the sphere-formation assay further showed that strengthened LINC00908 expression weakened the sphere-formation ability of PCa cells (Figure 1H). Meanwhile, we also conducted a limiting dilution assay to detect the secondary tumor sphere-formation capacity of LNCAP and PC3 cells. Results exhibited that many fewer secondary spheres were formed in the LINC00908 up-regulation group in comparison to the control group (Figure 1I). All aforementioned findings suggested that LINC00908 suppressed PCa cell stemness. To further test the role of LINC00908 in PCa development, in vivo assays were implemented. We revealed that PCa cells with LINC00908 overexpression formed smaller and lighter xenografts than the control group (Figure 1J–L). Altogether, these results suggested that LINC00908 hinders cell stemness and tumorigenesis in PCa.
2.2 LINC00908 Down-Regulation Is Mediated by the HDAC2-p300-YY1 Transcription Complex
We next probed the molecular mechanism of LINC00908 down-regulation in PCa cells. Transcriptional regulation is crucial for the expression of genes [25]. Hence, we utilized UCSC (http://genome.ucsc.edu) and JASPAR (http://jaspar.genereg.net) websites to find the transcription factors of LINC00908. As revealed in Figure 2A,B, YY1 was predicted to be the transcription factor of LINC00908 and possess 2 binding sites within the LINC00908 promoter. Meanwhile, based on docking models for HDOCK and Rosetta software, YY1 could interact with LINC00908 with low free energy in predicted binding sites (Figure 2C). Furthermore, YY1 functions in cancer cell stemness and prognosis for PRAD (Figure S2A–C). To confirm the effectiveness of this prediction, the impact of these two sites on the luciferase activity of the LINC00908 promoter was evaluated by HEK293T and PC3 cells after YY1 up-regulation. The data indicated that the luciferase activity of the wild-type LINC00908 promoter was significantly reduced when YY1 was increased, and site 1 or site 2 mutation partly counteracted such reduction, while YY1 up-regulation had no influence on the activity of the two sites-mutated LINC00908 promoter (Figure 2D). Meanwhile, ChIP assays further validated the combination between the LINC00908 promoter and YY1 at both sites (Figure 2E).
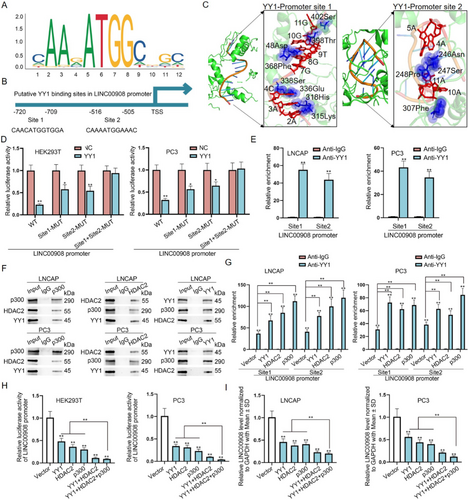
It has been affirmed that HDAC2 has the capacity to interact physically with YY1, p300 can interact with YY1 directly, and the formed HDAC2-YY1-p300 complex regulates gene transcription activity [26]. Therefore, we suspected that LINC00908 down-regulation was induced by the HDAC2-p300-YY1 complex. To confirm the existence of the HDAC2-p300-YY1 complex in PCa cells, we used anti-p300 and anti-HDAC2 to perform Co-IP assays. The results displayed that p300, HDAC2, and YY1 could interact with each other (Figure 2F). Through ChIP assays, we found that the overexpression of YY1, HDAC2, or p300 could increase the enrichment of YY1 in the LINC00908 promoter at both sites (Figure 2G). Afterward, it was revealed that the luciferase activity of the LINC00908 promoter, along with LINC00908 expression, was significantly reduced after the up-regulation of either YY1, HDAC2, or p300 and was further lowered under their co-overexpression (Figure 2H,I). Collectively, LINC00908 down-regulation in PCa cells is transcriptionally mediated by the HDAC2-p300-YY1 complex.
2.3 LINC00908 Inactivates the Wnt Pathway by Promoting the Ubiquitination and Degradation of β-Catenin
LncRNAs could exert functions on cancer progression via modulating some pathways [27]. Here, we first detected the relation of LINC00908 to several classical signaling pathways, including Notch, NF-κB, JAK/STAT3, Wnt, PI3K/AKT, NRF2, MAPK/JNK, Hedgehog, and MAPK/ERK pathways. Figure 3A showed that the LINC00908 overexpression led to a remarkable decline in the activity of the Wnt signaling pathway. β-catenin is a typically key mediator of the Wnt signaling pathway and functions in the nucleus eventually [28]. Thus, we performed immunofluorescence staining to assess the impact of LINC00908 on β-catenin expression. Western blotting analyses manifested that the levels of total and nuclear β-catenin declined after LINC00908 up-regulation, resulting in reduced levels of targeted factors including c-Myc, CCND1, SOX2, and SOX4 (Figure 3B, Figure S3A). Likewise, we observed that LINC00908 up-regulation weakened the intensity of β-catenin in the cytoplasm as well as nucleus of LNCAP and PC3 cells (Figure 3C).

Interestingly, the inhibition of LINC00908 on β-catenin intensity seemed to be dose-dependent (Figure S3B,C). The results hinted that LINC00908 might directly affect β-catenin expression in PCa cells. However, we found that LINC00908 could not modulate the mRNA level of CTNNB1 (β-catenin) (Figure 3D). Hence, we additionally assessed the impact of LINC00908 on the level of protein of β-catenin in LNCAP and PC3 cells. It manifested that after treatment utilizing the inhibitor for protein synthesis cycloheximide (CHX), the degradation of β-catenin sped up when LINC00908 was up-regulated (Figure 3E). Additionally, the treatment of the proteasome inhibitor MG132 could attenuate the suppressive effect of up-regulated LINC00908 on β-catenin protein (Figure 3F). We further implemented ubiquitination assays and found the level of ubiquitination of β-catenin protein was elevated when LINC00908 was overexpressed (Figure 3G). Taken together, LINC00908 destabilizes β-catenin to inactivate the Wnt pathway via promoting its ubiquitination and degradation.
2.4 LINC00908 Increases GSK3B Expression to Promote β-Catenin Degradation via Sponging miR-3179
It is well-known that β-catenin degradation is closely related to a multiprotein “destruction complex” formed by Axin, APC and GSK3B [29]. Hence, we employed RT-qPCR to detect the impact of LINC00908 on these three genes. We found that LINC00908 up-regulation significantly enhanced the expression of GSK3B, whereas it barely affected that of APC or AXIN (Figure 4A). LncRNAs exhibit the capacity to modulate GSK3B expression by acting as endogenous sponges of microRNAs (miRNAs) [30]. Herein, we also conjectured that LINC00908 might affect GSK3B expression by serving as a competing endogenous RNA (ceRNA) to sponge miRNAs. Therefore, we performed Ago2-RIP assays since Ago2 is the fundamental component of the miRNA-triggered silencing complex [31]. Data inferred that both LINC00908 and GSK3B were remarkably immunoprecipitated by anti-Ago2 (Figure 4B), suggesting the potential existence of a ceRNA network involving LINC00908 and GSK3B in PCa cells.
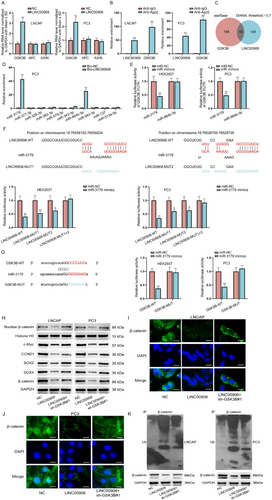
Then, potential miRNAs that combined with both LINC00908 and GSK3B were identified by the starBase (http://starbase.sysu.edu.cn) and DIANA (http://carolina.imis.athena-innovation.gr) databases. Ten miRNAs were predicted (Figure 4C). We then performed an RNA pull-down assay, discovering that miR-3179 and miR-664b-3p were only significantly pulled down by Bio-LINC00908 (Figure 4D). Intriguingly, through luciferase reporter assays, we confirmed that only miR-3179 overexpression caused remarkable attenuation in the luciferase activity of GSK3B 3′UTR (Figure 4E). We projected two binding sites between LINC00908 and miR-3179 via the DIANA database, and luciferase reporter assays further validated the effectiveness of both sites, as the activities of LINC00908-WT, LINC00908-MUT1, and LINC00908-MUT2 obviously declined while that of LINC00908-MUT1 + 2 was not changed when miR-3179 expression was elevated (Figure 4F). Similarly, the binding sequences between GSK3B and miR-3179 were predicted by the starBase database. It was then unveiled that the luciferase activity of GSK3B-WT was obviously inhibited when miR-3179 was under overexpression in HEK293T and PC3 cells (Figure 4G). Taken together, LINC00908 regulates GSK3B expression via competitively interacting with miR-3179.
To assess whether LINC00908 regulated GSK3B to affect PCa cell stemness and the Wnt signaling pathway, we then performed rescue experiments. Through Western blotting analysis, we found that GSK3B silence could partly reverse the inhibited effects of LINC00908 overexpression on the levels of nuclear β-catenin, Wnt pathway target genes (c-Myc, CCND1), and stemness-associated factors (SOX2 and SOX4) (Figure 4H). Meanwhile, the reduced staining of β-catenin mediated by LINC00908 up-regulation was partially restored after co-transfection of sh-GSK3B#1 (Figure 4I,J). In addition, the enhanced ubiquitination of β-catenin protein caused by LINC00908 up-regulation was moderately offset when GSK3B was down-regulated simultaneously (Figure 4K). All these data indicate that LINC00908 partly depends on GSK3B to promote β-catenin ubiquitination and inactivate the Wnt pathway.
2.5 LINC00908 Inactivates the Wnt Signaling Pathway to Suppress PCa Cell Stemness via Regulating FBXW2 and GSK3B
Next, we further explored another potential mechanism through which LINC00908 regulated β-catenin ubiquitination and degradation. We used RT-qPCR to analyze the influences of LINC00908 overexpression on the mRNA levels of some factors that have been reported to modulate the ubiquitination and degradation of β-catenin. As illustrated in Figure 5A, a significant up-regulation of FBXW2 was found in LNCAP and PC3 cells after LINC00908 overexpression. Furthermore, Western blotting analysis confirmed that LINC00908 up-regulation also enhanced the protein level of FBXW2 (Figure 5B, Figure S4A–C). To determine the regulatory patterns of LINC00908 on FBXW2 expression, we executed subcellular fractionation in conjunction with FISH experiments to identify the cellular location of LINC00908 in PCa cells. Results indicated that LINC00908 was principally distributed in the cytoplasm (Figure 5C,D), mirroring the post-transcriptional regulation of LINC00908 on FBXW2 expression.
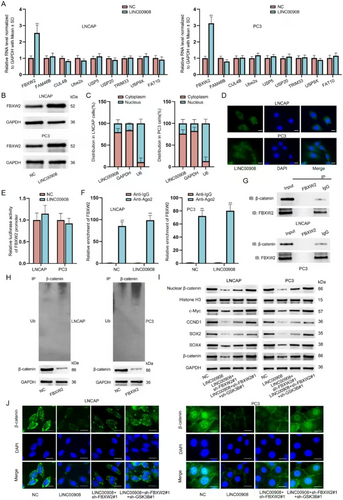
We excluded the transcriptional modulation of LINC00908 on FBXW2 because LINC00908 had no impact on the luciferase activity of the FBXW2 promoter (Figure 5E). CeRNA regulatory network is a common post-transcriptional regulation, and lncRNAs have been reported to regulate gene expression via the ceRNA model [32]. Thus, we surmised that LINC00908 might serve as a ceRNA to affect FBXW2 expression in PCa cells. Intriguingly, we found that LINC00908 up-regulation could not affect the combination between FBXW2 and Ago2 (Figure 5F), which eliminated the ceRNA regulatory network between LINC00908 and FBXW2. To test whether LINC00908 regulated FBXW2 to affect β-catenin ubiquitination and degradation, we first executed Co-IP assays to validate the combination between FBXW2 and β-catenin. As shown in Figure 5G, FBXW2 and β-catenin interacted with each other. We then up-regulated FBXW2 expression in LNCAP and PC3 cells (Figure S4D) and discovered that FBXW2 overexpression heightened the level of ubiquitination of β-catenin (Figure 5H). All these results proved that FBXW2 also engaged in the process of LINC00908-regulated β-catenin ubiquitination and degradation in PCa cells.
Thereafter, we performed rescue assays to validate whether GSK3B and FBXW2 were fully responsible for the effect of LINC00908 on PCa cell stemness and the Wnt signaling pathway. Based on Western blotting analysis results, we ascertained that the lessened level of nuclear β-catenin, Wnt pathway target genes (c-Myc, CCND1) and stemness-related factors (SOX2 and SOX4) caused by LINC00908 overexpression was partly restored after co-transfection of sh-FBXW2#1 and was totally reversed after co-transfection of sh-FBXW2#1 and sh-GSK3B#1 (Figure 5I). Similarly, FBXW2 silence partially offset the suppression of LINC00908 up-regulation on β-catenin, while the knockdown of FBXW2 and GSK3B completely counteracted the effect caused by LINC00908 overexpression (Figure 5J). Taken together, LINC00908 inactivates the Wnt signaling pathway to suppress PCa cell stemness via regulating FBXW2 and GSK3B.
2.6 LINC00908 Recruits DDX3X to Stabilize FBXW2 mRNA in PCa Cells
We further investigated the possible mechanism of LINC00908 in regulating FBXW2 expression. Here, we confirmed that LINC00908 could not function as a ceRNA to affect FBXW2 expression; thus, we guessed that LINC00908 might interact with certain RBPs to regulate FBXW2. We executed RNA pull-down assays to discover the protein partner of LINC00908. One specific band emerging on the electrophoretic gel at approximately 75 kDa was noticed in the biotin-labeled LINC00908 group rather than in the biotin-labeled LINC00908 Antisense group. The gel was then analyzed using mass spectrometry, and we finally identified DDX3X as an LINC00908-interacting protein in PCa cells (Figure 6A). Meanwhile, DDX3X expression was positively correlated with LINC00908 expression, and it also plays an important role in tumor progression for PRAD (Figure S5A–D). Moreover, the combination between LINC00908 and DDX3X, as well as the binding of DDX3X to FBXW2 in both LNCAP and PC3 cells, was verified via RNA pull-down and RIP assays (Figure 6B–E). Furthermore, based on catRAPID, AlphaFold2, and Rosetta software, LINC00908 was found that LINC00908 could interact with DDX3X (Figure 6F–H).
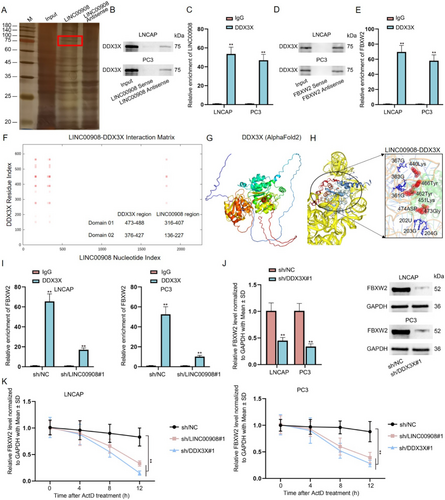
Subsequently, we planned to detect the influence of LINC00908 on the interaction between FBXW2 and DDX3X. It was proved that the loss of LINC00908 markedly hampered the binding of DDX3X to FBXW2 mRNA (Figure S5E, Figure 6I). Furthermore, we down-regulated the expression of DDX3X and found that DDX3X deficiency inhibited the mRNA and protein levels of FBXW2 (Figure S5F, Figure 6J). Additionally, we treated LNCAP and PC3 cells with actinomycin D (ActD) to inhibit the synthesis of FBXW2 mRNA and found that interference of LINC00908 or DDX3X weakened the stability of FBXW2 mRNA (Figure 6K). Collectively, LINC00908 recruits DDX3X to stabilize FBXW2 mRNA in PCa cells.
2.7 LINC00908 Represses PCa Cell Stemness via Up-Regulating FBXW2 and GSK3B
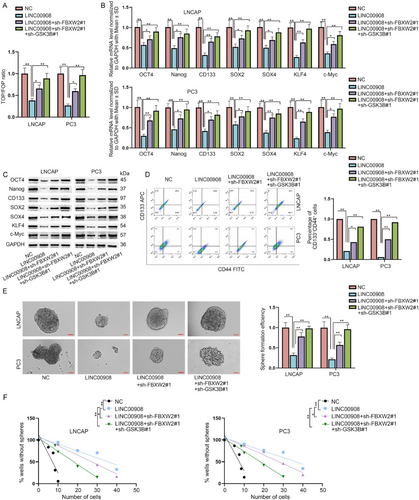
To further measure the effects of LINC00908/FBXW2/GSK3B on the Wnt pathway and PCa cell stemness, we conducted the following rescue assays. First of all, data from TOP/FOP assays evidenced that the inhibition of LINC00908 on Wnt activity was partially reversed by FBXW2 knockdown and was completely countervailed by co-inhibition of FBXW2 and GSK3B (Figure 7A). Consistent with this, reduced mRNA and protein levels of stemness-related factors caused by LINC00908 overexpression were partly restored after co-transfection of sh-FBXW2#1 and were completely recovered after co-transfection of sh-FBXW2#1 + sh-GSK3B#1 (Figure 7B,C). Meanwhile, the lessened percentage of CD44+CD133+ cells mediated by LINC00908 up-regulation was partly reversed after FBXW2 silence and was thoroughly recovered when FBXW2 and GSK3B were down-regulated simultaneously (Figure 7D). Additionally, FBXW2 deletion moderately offset the restraining effects of LINC00908 overexpression on primary and secondary sphere formation, while joint down-regulation of FBXW2 and GSK3B could fully neutralize such effects (Figure 7E,F). In conclusion, LINC00908 represses PCa cell stemness by inactivating the FBXW2/GSK3B-regulated Wnt pathway.
3 Discussion
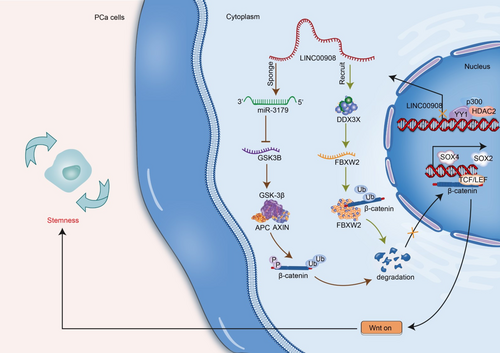
In our study, we identified that LINC00908 down-regulation in PCa was transcriptionally mediated by the HDAC2-p300-YY1 complex. Significantly, LINC00908 inhibited PCa cell stemness by regulating miR-3179/GSK3B and DDX3X/FBXW2 pathways to promote the ubiquitination and degradation of β-catenin and inactivate the Wnt pathway. The findings implied that LINC00908 might be a possible target for PCa therapy (Figure 8).
In many tumors, cell stemness is proven to function in sustaining tumorigenesis and progression [33]. Current literature has demonstrated the regulatory function of LINC00908 in several human cancers. In particular, LINC00908 is responsible for hepatocellular carcinoma progression by increasing the stability of SOX-4 [34]. LINC00908 facilitates colorectal cancer cell proliferation via modulating the miR-143-3p/KLF5 axis [35]. Our research found that LINC00908 was significantly low expressed in PCa cells. Overexpression of LINC00908 repressed PCa cell stemness and in vivo tumor growth, which indicated the tumor suppressive property of LINC00908 in PCa.
Moreover, we affirmed that YY1 was the transcription factor of LINC00908 in PCa cells. YY1 is a zinc finger transcription factor that has been linked to the development of distinct malignancies, including PCa [36]. YY1 could interact with p300 and HDAC3 as well as HDAC6 to repress gene transcription [37, 38]. Consistently, our study revealed the interaction among YY1, p300, and HDAC2 in PCa cells, and that LINC00908 was transcriptionally repressed through the YY1/p300/HDAC2 complex.
Importantly, our study disclosed that LINC00908 inactivated the Wnt pathway in PCa cells. Many studies have uncovered that lncRNAs regulate the Wnt pathway to impede PCa progression [39]. β-catenin is a key molecule of the Wnt pathway, since it can translocate into the nucleus to modulate gene expression upon Wnt stimulation [40]. Currently, we found that LINC00908 facilitated the ubiquitination and degradation of β-catenin to reduce total and nuclear β-catenin levels, thereby inactivating the Wnt pathway in PCa cells. As we know, β-catenin is degraded by a multiprotein “destruction complex” containing Axin, APC, and GSK3B [41]. Here, we confirmed that LINC00908 positively regulated GSK3B expression via acting as a ceRNA to sponge miR-3179. A study proposed by Alfredo I Sagredo et al. has pointed out the regulatory relation between GSK3B and β-catenin activity in PCa cells [42]. Similarly, the present work verified that LINC00908 up-regulated GSK3B expression to accelerate the ubiquitination and degradation of β-catenin and thereby inactivated the Wnt pathway. However, LINC00908 depended partly on GSK3B to affect β-catenin ubiquitination and Wnt inactivation.
Fortunately, we further uncovered that LINC00908 strengthened FBXW2 stability to facilitate the ubiquitination and degradation of β-catenin. FBXW2 is a substrate recognition receptor of E3 ubiquitin ligase SCF (a complex of FBXW2, SKP1, and cullin-1) [43]. FBXW2 could inhibit cancer progression by promoting β-catenin ubiquitination and degradation [44], which is in line with our study. More importantly, our study further testified that LINC00908 recruited DDX3X to stabilize FBXW2 mRNA. Taken together, our study confirmed that LINC00908 up-regulated FBXW2 and GSK3B to inactivate the Wnt/β-catenin pathway, consequently inhibiting PCa cell stemness.
Briefly, our study elucidates the suppressive functions of LINC00908 in PCa cell stemness, which might offer a meaningful revelation for exploring new and effective PCa treatment. However, the absence of clinical data is a major drawback of the present work, and we will perfect the work in the future.
4 Material and Methods
4.1 Cell Culture
PCa cells (DU145: CLCV 0105, 22RV1: CLCV 1045, VCAP: CLCV 2235, LNCAP: CLCV 0395 and PC3: CLCV 0035), human normal prostatic epithelial cells (RWPE-1) and human embryonic kidney 293 T (HEK293T) cells were supplied by ATCC (Manassas, VA, USA). DU145 cells grew in Eagle's Minimum Essential Medium. 22RV1, LNCAP and PC3 cells were cultured in RPMI-1640 Medium. VCAP and HEK293T cells were cultivated in Dulbecco's Modified Eagle's Medium. RWPE-1 cells grew in Keratinocyte Serum Free Medium (Gibco) mixed with 0.05 mg/mL BPE and 5 ng/mL EGF. All cell culture media contained 10% fetal bovine serum (FBS), and cells were sustained at 37°C with 5% CO2.
Short tandem repeating (STR) profiling authenticated all cell lines for cell availability within the last 3 years before testing for mycoplasma contamination utilizing Mycoplasms Stain Assay Kit (Beyotime, China), and all the experiments were executed utilizing mycoplasma-free cells. The last time for a cell test was Feb 2023.
4.2 Reagents and Kits
Cycloheximide (CHX; 50 μg/mL) and actinomycin D (ActD; 4 μM) were purchased from Sigma-Aldrich (St. Louis, MO, USA), while Selleck Chemicals (Houston, TX, USA) supplied MG132 (25 μM).
For the activity detection of several signaling pathways, Notch1/CSL Reporter Kit (amsbio, Madrid, Spain), NF-κB (p65) Transcription Factor Assay Kit (Cayman, Cayman, England), STAT3 Transcription Factor Assay Kit (Cayman), TCF/LEF Reporter kit (for Wnt pathway; amsbio), AKT Activity Assay Kit (Biovision, San Francisco, CA, USA), NRF2 Transcription Factor Assay Kit (Cayman), Human sonic hedgehog ELISA kit (amsbio), JNK Activity Assay kit (Abcam), and SRE Reporter Kit (for MAPK/ERK Signaling Pathway; amsbio) were applied.
4.3 Cell Transfection and Virus Infection
The overexpression vectors for LINC00908, YY1, HDAC2, p300, and FBXW2 and the respective negative control (NC), miR-3179 mimics and miR-NC, sh-GSK3B#1 and sh-NC, sh-FBXW2#1 and sh-NC, shRNAs targeting LINC00908 (sh/LINC00908#1 and sh/LINC00908#2) and sh/NC, shRNAs targeting DDX3X (sh/DDX3X#1 and sh/DDX3X#2) and sh/NC were cloned utilizing Genepharma. The transfection of miR-3179 mimics and miR-NC was carried out via Lipofectamine 3000 (Invitrogen, USA) as per the manufacturer's protocol. To trigger gene overexpression, corresponding cDNA was cloned into GV502 lentiviral vectors (Genechem), accompanied by the transfection of these lentiviral particles into indicated PCa cells. The transfection of shRNA entailed the utilization of transduction particles of GV248 (Genechem), which carried the indicated shRNAs and were deployed for transfecting both LNCAP and PC3 cells. Cells with stable transfections were selected by puromycin (1 μg/mL) and then kept for next use.
4.4 Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
TRIzol reagent (Invitrogen) extracted total RNA from PCa cells. PrimeScript RT master mix (Takara, Japan) and SYBR Green PCR Master Mix (Applied Biosystems, USA) executed reverse transcription and qPCR, respectively. U6 or Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the 2−ΔΔCt method acted as an internal reference and determined gene expression, respectively. Analyses were triplicated.
4.5 Western Blotting Analysis
Extraction of total protein from PCa cells was realized utilizing RIPA Lysis buffer (Thermo Fisher, USA). Upon confirming protein concentration with a BCA Protein Assay Kit (Beyotime, USA), proteins were obtained using 10% SDS-PAGE and subjected to PVDF membranes (Millipore, USA). It was accompanied by sealing the membranes with 5% defatted milk and then hatched with the primary antibodies containing anti-OCT4 (Abcam, 1/1000), anti-Nanog (Abcam, 1/1000), anti-CD133 (Abcam, 1/1000), anti-SOX2 (Abcam, 1/1000), anti-SOX4 (Abcam, 1 μg/mL), anti-KLF4 (Abcam, 1/1000), anti-nuclear β-catenin (Abcam, 1/4000), anti-c-Myc (Abcam, 1/1000), anti-CCND1 (Abcam, 1/200), anti-β-catenin (Abcam, 1/4000), anti-histone H3 (Abcam, 1/1000), anti-FBXW2 (Abcam, 1/500), anti-DDX3X (Abcam, 1/1000) and anti-GAPDH (Abcam, 1/1000) overnight at 4°C. Subsequent to washing the membranes, they were hatched with secondary antibodies at room temperature for 1 h. The blots were measured utilizing an ECL Western blotting substrate, and the experiments were triplicated.
4.6 Flow Cytometry Analysis
The CD44+CD133+ cell proportion was measured utilizing a flow cytometer FACSCalibur (BD Biosciences). Cells (1 × 106) were washed and then incubated with 10 μL of CD44 and CD133 antibody. Next, the cells were washed and centrifuged, accompanied by analysis utilizing the aforementioned flow cytometer. Experiments were triplicated.
4.7 Sphere-Formation and Limiting Dilution Assays
Cells grew in 6-well ultra-low attachment plates (Corning, USA) containing serum-free medium treated with FGF, EGF, N2, and B27 (Gibco). After incubation for 2 weeks, the formed spheres were counted as well as imaged using a light microscope.
For limiting dilution assay, the stem cells obtained from the above spheres were collected and dissociated into single cells, and then the indicated number of cells was planted into each well of 96-well plates. Nine days later, wells with no secondary tumor spheres were counted for each group. Experiments were conducted in triplicate.
4.8 In Vivo Tumorigenesis Experiment
For the in vivo xenograft assay, mice were classified into two groups randomly. Afterward, about 1 × 107 of PC3 cells with LINC00908 overexpression in conjunction with those without were subcutaneously inoculated into the left dorsal of male nude mice (Slac Laboratories, Shanghai, China; 4 weeks old, n = 6), respectively. The xenograft sizes were documented at a frequency of 4 days. Upon the fourth week, the animals were sacrificed, and the tumors were excised and weighed. The animal experiments received approval from the Ethics Committee of the First Affiliated Hospital of Bengbu Medical College.
4.9 Luciferase Reporter Assay
For gene transcription analyses, cells were transfected together with specified transfection plasmids and the pGL3-basic reporter vectors (Promega, Madison, WI, USA) containing wild type (WT) or mutant type (MUT) LINC00908 promoter or FBXW2 promoter. Two days later, the luciferase activity of specified reporters was monitored via a dual luciferase reporter assay kit (Promega).
In addition, LINC00908 or GSK3B 3′untranslated regions (3′UTR) fragments containing miR-3179 WT or MUT binding sites were cloned into pmirGLO luciferase reporter vectors (Promega), and subsequently transfected together with miR-3179 mimics or NC mimics into HEK293T and PC3 cells. At the end of 48 h of transfection, the activities of luciferase were detected using a dual luciferase reporter assay kit (Promega). Experiments were conducted in triplicate.
4.10 Chromatin Immunoprecipitation (ChIP)
ChIP kit (Beyotime, USA) performed ChIP assays as per the manufacturer's suggestions. Cells were processed with cross-linking and sonication to obtain genomic DNA fragments. The derived DNA fragments were immunoprecipitated with antibodies against YY1 or normal mouse IgG. Then, qPCR detected the immunoprecipitated DNAs. The experiment was triplicated.
4.11 Co-Immunoprecipitation (Co-IP)
Cells were lysed and collected for immunoprecipitation using anti-p300, anti-HDAC2, anti-β-catenin, anti-FBXW2, or anti-IgG (Abcam) for 1 h. Afterward, the antibody-protein complex was hatched with protein A/G agarose beads, and the co-immunoprecipitated protein complex was then examined by Western blotting analysis. The assay was triplicated.
4.12 Immunofluorescence Staining
Cells were planted into 6-well plates and cultured on coverslips that were deemed sterilized, fixed, and permeabilized. After blocking, cells were incubated with primary antibodies against β-catenin at 4°C overnight, accompanied by further processing with fluorochrome-labeled secondary antibodies for 1 h. DAPI dyed cell nuclei, and the images were captured utilizing fluorescence microscopy. The experiment was triplicated.
4.13 Ubiquitination Assay
After cell lysis and centrifugation, the cells underwent treatment with β-catenin-specific antibody and protein A/G agarose beads overnight at 4°C. Upon rinsing, the precipitated proteins were boiled and then processed with SDS-PAGE. A monoclonal antibody against ubiquitin was utilized in ascertaining the level of ubiquitination of β-catenin. The assay was triplicated.
4.14 RNA Binding Protein Immunoprecipitation (RIP)
RIP was conducted with a RIP Kit (Millipore). Lysates were incubated with Ago2 antibody, DDX3X antibody, or normal mouse IgG antibody. Protein A/G agarose beads immunoprecipitated the RNA-protein complexes. RNAs were purified and then analyzed via RT-qPCR. The assay was performed in triplicate.
4.15 RNA Pull-Down Assay
Biotin-labeled LINC00908 or FBXW2 full-length sense and antisense were synthesized. Biotin-labeled RNAs were severally incubated with cell lysates, and the eluted proteins were purified and analyzed by either Western blotting analysis or silver staining. This assay was triplicated.
4.16 Subcellular Fractionation
The isolation of nuclear as well as cytosolic fractions was conducted via a PARIS Kit (Invitrogen) conforming to the specifications. The RNA levels of LINC00908, GAPDH, and U6 in the nuclei and cytoplasm were evaluated by RT-qPCR. Experiments were triplicated.
4.17 Fluorescent In Situ Hybridization (FISH)
The digoxin-labeled LINC00908 probe purchased from Invitrogen was applied in the FISH assay. Cells were fixed, and then incubated with the digoxin-labeled LINC00908 probe in the hybridization mixture all night. Then, the slides were washed, blocked, and incubated with biotin-conjugated anti-digoxin antibody, followed by further processing with SABC-FITC for half an hour at 37°C. Cell nuclei were dyed with DAPI, and the fluorescence of LINC00908 was visualized via a fluorescence microscope. The experiment was performed in triplicate.
4.18 TOP/FOP Flash Assay
For this experiment, TOP/FOP-Flash (Genechem) was co-transfected into indicated cells which were previously transfected with LINC00908 overexpression plasmids, sh-FBXW2#1, sh-GSK3B#1, or corresponding control vector as needed. The Renilla reniformis (Promega) was used as the normalized control for TOP/FOP flash values. The TOP/FOP ratio was measured to analyze Wnt activity. The assay was performed in triplicate.
4.19 Protein-RNA Interaction Analysis
The three-dimensional structures of proteins were predicted by the AlphaFold 2 web server. The catRAPID was used to compute the overall RNA-binding regions in proteins and the corresponding regions in RNA. Then, the secondary and three-dimensional structures of RNA were predicted by RNAfold and HDOCK software. Based on energy scores, the complex structure with the lowest energy was chosen to optimize by Rosetta software. The interaction sites in the complex structure were analyzed based on spatial coordinates and biochemical properties.
4.20 Bioinformatics Analysis for PRAD
The GSE70769 data was downloaded from the GEO database. The significance difference in lncRNA in PRAD was calculated when compared to adjacent normal tissues by the DEGseq algorithm. HR (hazard ratio) values with a 95% confidence interval were calculated by the coxph function in R software based on the TCGA database. Then, the probability of differences in relapse-free survival was ascertained by the Kaplan–Meier method with the log-rank test for significance. We further probed the correlation between the expression of LINC00908/YY1 and the stemness index of the tissue samples containing mRNA expression-based stemness scores (RNAss) across multiple cancer types using the Spearman's correlation test. Furthermore, correlation coefficient values for two protein expressions were calculated by Pearson. Both pathology images for proteins in PRAD patients and subcellular location images for proteins were downloaded from the HPA (human protein atlas) web server.
4.21 Statistical Analyses
For each independent experiment, three bio-repeats were run in technical triplicate. Collected data were analyzed using GraphPad Prism 6 software, and presented as mean ± standard deviation (SD). Differences were assessed using Student's t-test, one-way or two-way analysis of variance (ANOVA). Tukey or Dunnett's test was used for back testing. A p < 0.05 was defined as statistically significant. The consistent data collected from each experiment were presented ultimately. Data which could not form a normal distribution and negative results were excluded. Criteria for data exclusion were determined in advance. The group allocation and outcome assessment were conducted based on random selection. Meanwhile, biological experiments were conducted in triplicate for statistical analyses.
Author Contributions
Han Guan: conceptualization (equal), data curation (equal), formal analysis (equal), funding acquisition (equal), investigation (equal), methodology (equal), resources (equal), supervision (equal), validation (equal), visualization (equal), writing – original draft (equal), writing – review and editing (equal). Qiang Hu: conceptualization (equal), data curation (equal), formal analysis (equal), resources (equal), supervision (equal), validation (equal), writing – original draft (equal). Lilin Wan: formal analysis (equal), visualization (equal), writing – original draft (equal). Can Wang: formal analysis (equal), investigation (equal), methodology (equal), resources (equal), validation (equal). Yifeng Xue: investigation (equal), resources (equal), supervision (equal). Ninghan Feng: funding acquisition (equal), writing – review and editing (equal). Ming Chen: funding acquisition (equal), writing – review and editing (equal). Chenggui Zhao: funding acquisition (equal), supervision (equal). Zonghao You: conceptualization (equal), data curation (equal), formal analysis (equal), funding acquisition (equal), investigation (equal), methodology (equal), resources (equal), validation (equal), visualization (equal), writing – original draft (equal), writing – review and editing (equal).
Acknowledgments
We thank all individuals and teams who have participated in or contributed to our work.
Ethics Statement
All animal experiments were approved by the First Affiliated Hospital of Bengbu Medical College and Zhongda Hospital Institutional Animal Care and Use Committee.
Consent
All authors approved the manuscript for submission and gave their consent for publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.