Highly expressed carbohydrate sulfotransferase 11 correlates with unfavorable prognosis and immune evasion of hepatocellular carcinoma
Abstract
Despite great advance has been made in multi-modality treatments for HCC patients, the effectiveness is far from satisfactory with worse survival outcome, which may be partly explainable by the anti-tumor deficiency of the immune system. It is necessary to clarify the molecular mechanism of HCC immunodeficiency. Here, we demonstrated that carbohydrate sulfotransferase 11 (CHST11) was upregulated in HCC and related to advanced TNM stage. HCC patients with TP53 mutation showed higher CHST11 expression. Survival analysis revealed that CHST11 was an independent prognostic biomarker in HCC. Cellular functional experiments indicated that knockdown of CHST11 in HCC inhibited cell proliferation and metastasis. Gene functional enrichment analyses indicated that CHST11 modulated pathways related to tumor growth, metastasis and immune regulation. Continuative immune-related analyses revealed that CHST11 expression facilitated Tregs infiltration in HCC and promoted the expression of checkpoints PD-L1/PD-1, resulting in the immunosuppression of HCC. Targeting CHST11 may inhibit Tregs infiltration and enhance the antineoplastic effect of immune checkpoint inhibitors, which provides a novel insight into the combination immunotherapy with Treg-modulating agents and PD-L1/PD-1 inhibitors.
1 INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most frequently occurring malignances and ranks third in global tumor-related mortality, with approximately 780,000 deaths and 840,000 new cases annually.1 Despite great advance has been made in the field of diagnosis and multi-modality treatments, the effectiveness is far from satisfactory with poor prognosis for HCC patients, which is partly explained by the anti-tumor deficiency of host immune system.2
With the increasing recognition of the role of immune system in tumor growth, researchers have focused on the immunotherapy in cancer therapy. Tumor immunotherapy aims to active host immune system to fight against cancer cells,3 and has shown obvious antineoplastic activity in the treatment of malignant tumors.4-7 Due to the eximious efficacy, immunotherapy has been awarded the “Breakthrough of the Year” in 2013.8 Although immunotherapy has undoubtedly revolutionized treatment strategies for cancer patients, there is still a percentage of cases that fail to respond to immunotherapy, which may be caused by the complexity and diversity of the host tumor microenvironment (TME).9 Immune cells in human body constantly surveil the cells around them. Once an abnormality is found, they will immediately activate the immune system to eliminate the abnormal cells, such as foreign pathogens, damaged cells and tumor cells.10 However, cancer cells can develop escape strategies to bypass the host immunosurveillance. An immunosuppressive TME exerts a key role in providing an immune-permissive environment for tumor growth, and regulatory T cells (Tregs) are important participants in the suppressive immune microenvironment, which lead to immunotherapy resistance.11 The core of this immunosuppressive environment for tumors is the activation of oncogenes and abnormal signaling pathways. Therefore, it is of great significance to clarify the key genes or signal pathways that regulate immunosuppressive TME in malignant tumors to improve the clinical outcome for tumor patients. Nevertheless, the molecular events that regulate immunosuppressive TME in HCC are not clear.
Carbohydrate sulfotransferase 11 (CHST11) encodes chondroitin 4-sulfotransferase 1, a key enzyme involved in the synthesis of chondroitin sulfate glycosaminoglycans (CS).12 As a vital member of the extracellular matrix (ECM), CS interacts with various proteins and cell surface-related ECM components in the TME to modulate tumor growth and metastasis.13 Given that limited evidence has illustrated the function and clinical significance of CHST11 in HCC, we conducted this study and integrated multiple evidences to investigate whether CHST11 is involved in HCC progression and immune regulation.
2 MATERIALS AND METHODS
2.1 Detection of CHST11 mRNA in HCC by in-house RT-qPCR
We collected 20 paired fresh-frozen HCC samples and adjacent liver samples from patients with tumor resection before any preoperative therapy for real-time quantitative polymerase chain reaction (RT-qPCR) analysis. The cases were diagnosed by senior pathologists.
RNA extraction, RNA reverse transcription and RT-qPCR experiment were executed with the manufactures' instruction. The primers for CHST11 (forward 5′-CACAAGCCGTAAGCGGAGG-3′; reverse 5′-CATGGGGTCGCTGTACTTCC-3′) and endogenous reference gene beta-action (forward 5′-TGGCACCCAGCACAATGAA-3′; reverse 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′) were purchased from Sangon Biotech and Takara, respectively. The CT value of each sample was detected and CHST11 expression was determined with the 2−ΔCT method. Paired t-test enables comparison of difference between groups. A p-value <0.05 indicates a statistical significance.
2.2 Validation of CHST11 mRNA in HCC by multi-center datasets
HCC-related gene microarrays and RNA-sequencing expression profiling were obtained from Gene Expression Omnibus (GEO)14 with following search strategy: (“hepatocellular carcinoma” OR “HCC” OR “liver cancer”) AND (gene OR mRNA). Strict criteria for dataset inclusion were established: (I) patients should be diagnosed with HCC, (II) processed or raw expression data should be provided, (III) the expression values of CHST11 should be detected and (IV) the number of samples in each group should be at least three. Accordingly, the exclusion standards were as follows: (I) research objects were not human beings, (II) the expression data of CHST11 in HCC were unavailable and (III) the samples were duplicated. In addition, high-throughput sequencing datasets were also collected from The Cancer Genome Atlas (TCGA)15 and International Cancer Genome Consortium (ICGC).16
The expression values of CHST11 in each included dataset were log2-transformed. The overall standard mean deviation (SMD) was determined with STATA 12.0 (Stata Corporation). SMD >0 with 95% confidence interval (CI) not containing zero indicates that CHST11 is highly expressed in HCC, SMD ≤0 with 95%CI not containing zero indicates that CHST11 is low expressed in HCC. Publication bias was assessed with Begg's and Egger's tests.
2.3 Clinical significances CHST11 mRNA in HCC
Clinicopathologic and survival information of HCC patients were collected from TCGA. Independent T-test was used to disclose the relation between CHST11 mRNA expression and clinicopathologic parameters (age, gender, pathology grading, TNM stage and TP53 mutation) of HCC. Univariate and multivariate COX analyses were applied to elucidate the prognosis prediction capability of CHST11 mRNA in HCC. The prognostic role of CHST11in HCC was corroborated by the Kaplan–Meier plotter.17
2.4 Expression of CHST11 protein in HCC
CHST11 protein expression was detected by immunohistochemistry (IHC) staining in 90 HCC and adjacent normal liver specimens, which were gained from the First Affiliated Hospital of Guangxi Medical University. The cases received no preoperative radio-chemotherapy and were diagnosed pathologically after surgery. The rabbit polyclonal antibody of CHST11 (1:100 dilution) were purchased from Abcam. The experiment was executed with the manufacturer's instruction.
CHST11 protein is mainly located in cytoplasm. Two pathologists independently assess the staining intensity and percentage of CHST11 protein in 10 consecutive and representative fields (×400 magnification microscope) for each slice. The expression of CHST11 protein was assessed with the immunoreactive scores (IRS).18 Sample was determined as CHST11 high expression (IRS >6) or CHST11 low expression (IRS ≤6). Mann–Whitney U non-parametric test was employed to compare the CHST11 protein expression between groups. A receiver operating characteristic (ROC) curve was generated to assess the capability of CHST11 protein to discriminate HCC samples from normal liver samples.
2.5 Clinical significances CHST11 protein in HCC
The clinical information of the 90 cases was presented in Table S1. Mann–Whitney U non-parametric test was used to analyze the relationship between CHST11 expression and clinicopathological parameters of age, gender, tumor size, pathology grading and Barcelona Clinic Liver Cancer (BCLC) stage.
2.6 In vitro experiments
2.6.1 Cell culture
HCC cells Huh7 and Hep3B were incubated in cell incubator at 37°C with DMEM medium containing 10% fetal bovine serum.
2.6.2 Cell transfection
CHST11 small interfere RNA (siRNA) lentiviruses were purchased from Genechem. Two CHST11 siRNA lentiviruses (si-81566 and si-81567) and control vector were transfected into Huh7 and Hep3B cells according to the manufacture’ instruction. RT-qPCR method was used to verify the knockdown efficiency.
2.6.3 Cell functional experiments
CCK8 test was executed to measure the effect of CHST11 silencing on HCC cell viability according to the manufactures' protocol. Wound-healing test was performed to evaluate the effect of CHST11 silencing on HCC cell migration according to the previous study.19
2.7 Collection of genes correlated to CHST11 in HCC
To obtain differently expressed genes (DEGs) in HCC, we collected data from TCGA and The Genotype-Tissue Expression (GTEx).20 DEGs were calculated by limma-voom in R package after removing of batch effect and identified with the threshold of |log2(fold change)| ≥ 1 and adjusted p < 0.05. Correlations of the DEGs and CHST11 in HCC were analyzed using Pearson correlation analysis. Genes with correlation coefficient >0.3 and p < 0.05 were regard as significantly correlated to CHST11.
2.8 Enrichment analyses
Gene Ontology analysis of biological process and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were executed with R package ClusterProfiler. KEGG pathway was visualized with pathview package in R. Gene-set enrichment analysis (GSEA) was conducted with GSEA 4.1.0 (BROAD Institute).
2.9 Tumor immune infiltration analyses
Immune and stromal scores of 371 HCC patients were computed with R package estimate based on data from TCGA, and the fractions of 22 infiltrating immune cells in individual HCC sample were assessed with CIBERSORT in R. Our study gauged the correlations between CHST11 expression and immune score, stromal score and immune cell subpopulations in patients with HCC. Considering that the CIBERSORT algorithm may suffer from statistical multicollinearity caused by the inclusion of highly correlated immune cell types, we employed the TIMER2 website21 to corroborate immunomodulatory role of CHST11 in HCC.
3 RESULTS
3.1 Expression of CHST11 mRNA in HCC
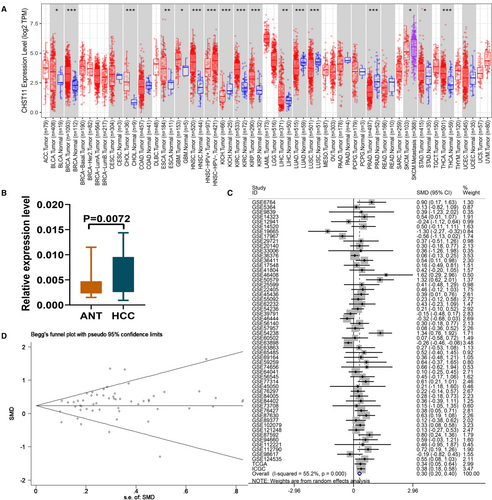
CHST11 mRNA expression in human cancers was first analyzed using the TCGA RNA-sequencing data. Increased expression of CHST11 mRNA was observed in bladder urothelial carcinoma, breast invasive carcinoma, cholangiocarcinoma, esophageal carcinoma, glioblastoma multiforme, head and neck squamous cell carcinoma, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, liver hepatocellular carcinoma, stomach adenocarcinoma and Thyroid carcinoma (Figure 1A). Consistently, we corroborated the elevated expression of CHST11 mRNA in HCC by RT-qPCR (p = 0.0072; Figure 1B). Simultaneously, we recruited 56 multi-center datasets involving in 3110 HCC samples and 2016 non-tumor samples from 12 countries around the world to further validate the expression of CHST11 mRNA in HCC. Figures S1-S3 presented the expression of CHST11 in the 56 datasets. Since the expression of CHST11 in individual datasets were not solid, we calculated the SMD based on the 56 datasets. A robust result showed that CHST11 was remarkably highly expressed in HCC tissues (SMD = 0.30, 95% CI = 0.20–0.40, p < 0.0001; Figure 1C), consistent with the RT-qPCR. No publication bias existed in this study (Begg's p = 0.682, Egger's p = 0.645; Figure 1D).
3.2 Clinical significances of CHST11 mRNA in HCC
CHST11 mRNA expression was related to TNM stage, but not to age, sex, or pathological grading in HCC. Patients with TNM stage III-IV exhibited higher CHST11 mRNA expression than those with TNM stage I-II (p = 0.0304; Figure 2A). Additionally, we assessed the association between CHST11 mRNA expression and TP53 mutation in 358 HCC samples, and found that CHST11 mRNA was highly expressed in mutated TP53 group than in wild-type TP53 group (p < 0.0001; Figure 2B).
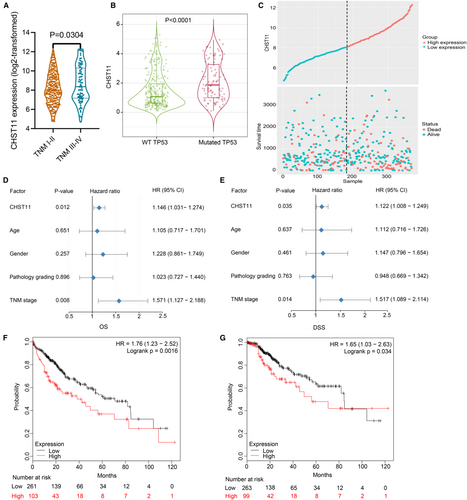
To assess the prognostic value of CHST11 in HCC, we collected 370 HCC patients with survival information from TCGA and divided them into high- and low-CHST11 expression groups according to the median expression value of CHST11 (Figure 2C). Univariate COX analysis revealed that increased CHST11 mRNA expression and advanced TNM stage in HCC predicted worse survival outcome (CHST11: hazard ratio [HR] = 1.146, 95% CI = 1.031–1.274, p = 0.012; TNM stage: HR = 1.571, 95% CI = 1.127–2.188, p = 0.008; Figure 2D). Multivariate COX analysis further revealed that CHST11 mRNA and TNM stage were independent prognostic biomarkers in HCC (CHST11: HR = 1.122, 95% CI = 1.008–1.249, p = 0.035; TNM stage: HR = 1.517, 95% CI = 1.089–2.114, p = 0.014; Figure 2E). Furthermore, data from the Kaplan–Meier plotter validated that HCC patients with higher expression level of CHST11 showed shorter overall survival (HR = 1.76, 95% CI = 1.23–2.52, p = 0.0016; Figure 2F) as well as disease-specific survival (HR = 1.65, 95% CI = 1.03–2.63, p = 0.034; Figure 2G).
3.3 Expression of CHST11 protein in HCC
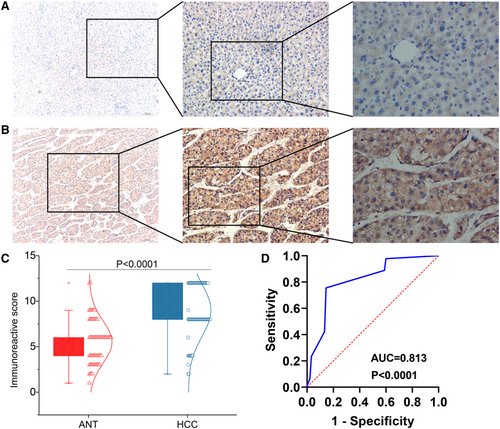
We scored CHST11 expression in the 90 HCC tissues and matched non-tumor tissues with IRS system. In HCC group, 77 samples showed high CHST11 expression and 13 samples showed low CHST11expression, while in adjacent non-tumor group, 21 samples showed high CHST11 expression and 69 samples showed low CHST11 expression. CHST11 protein expression was obviously increased in HCC specimens than in non-tumor specimens (Z = 67.901, p < 0.0001; Figure 3A-C and Figure S4). A ROC curve was generated with the area under the curve (AUC) value of 0.813 (95% CI = 0.749–0.877, p < 0.0001; Figure 3D), suggesting the capability of CHST11 to differentiate HCC specimens from non-cancerous liver specimens.
3.4 Relationship between CHST11 expression and clinicopathological parameters in HCC
The relationship between CHST11 expression and clinicopathological parameters (age, gender, pathology grading and BCLC stage) in HCC has been investigated. However, the results showed no relationship between CHST11 expression and these clinicopathological characteristics (Table S1).
3.5 Effects of silenced CHST11 on HCC cell biological behaviors
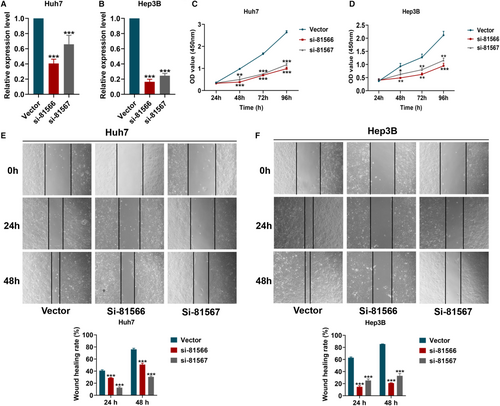
CHST11 expression was significantly reduced in Huh7 and Hep3B cells transfected with CHST11 silenced lentivirus (si-81566 and si-81567) (Figure 4A-B). As shown in Figure 4C-F, the cell proliferation and wound healing rates in si-81566 and si-81567 groups were remarkably lower than that in vector group.
3.6 Functional enrichment analysis of genes correlated to CHST11 in HCC
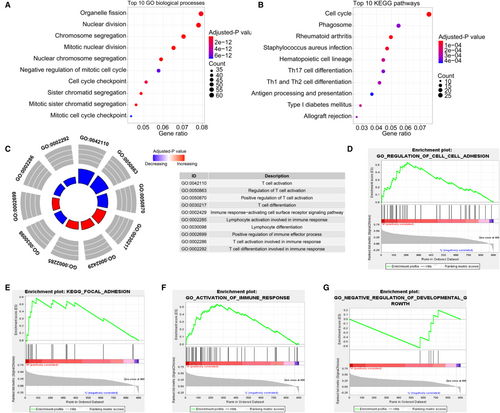
Totally, 4251 DEGs in HCC were obtained, of which 900 genes were significantly correlated with CHST11 (Figure S5). Functional enrichment analysis demonstrated that these genes participated in cell growth- and metastasis-related processes, such as “nuclear division”, “cell cycle checkpoint”, “cell cycle” and “extracellular matrix structural constituent” (Figure 5A-B). Interesting, we also found that the 900 genes involved in immune-related biological processes and pathways, such as “T cell activation”, “T cell differentiation” and “PD-L1 expression and PD-1 checkpoint pathway in cancer” (Figure 5C and Figure S6). Further GSEA analysis revealed that high expression of CHST11 positively regulated cell–cell adhesion (Figure 5D), focal adhesion (Figure 5E) and activation of immune response (Figure 5F), and low expression of CHST11 negatively regulated cell growth (Figure 5G).
3.7 Correlation of CHST11 expression and immune cell infiltration
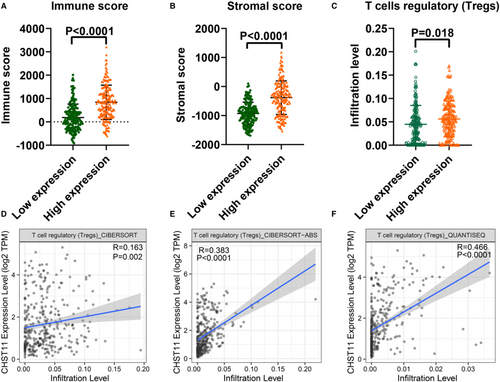
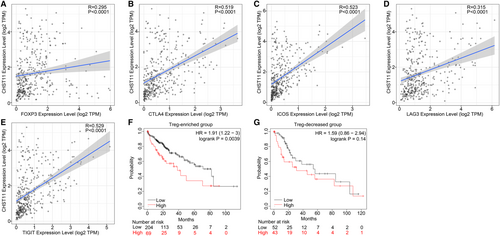
TME is a complex milieu of tumor epithelial cells, immune cells, and stromal cells. To elucidate the regulatory role of CHST11 in TME in HCC, we gauged the relationship between CHST11 expression and immune cells as well as stromal cells. The results indicated that patients with increased CHST11 expression had higher immune score (p < 0.0001) and stromal score (p < 0.0001) compared to patients with decreased CHST11 expression (Figure 6A,B), suggesting more immune and stromal cells in patients with high CHST11 expression. Furthermore, we assessed the correlation between CHST11 expression and immune cell infiltration with CIBERSORT algorithm, and found that the infiltration of Tregs was higher in high-CHST11 expression group than in low-CHST11 expression group (p = 0.018; Figure 6C). The positive correlation between CHST11 expression and Tregs infiltration were corroborated by TIMER2 with algorithms of CIBERSORT (R = 0.163, p = 0.002; Figure 6D), CIBERSORT-ABS (R = 0.383, p < 0.0001; Figure 6E), and Quantiseq (R = 0.466, p < 0.0001; Figure 6F). Simultaneously, we demonstrated that CHST11 expression was closely related to Treg cell intracellular and extracellular markers (FOXP3, CTLA4, ICOS, LAG3 and TIGIT) (Figure 7A-E), which were involved in the activation or differentiation of Tregs. We divided HCC patients into Treg-enriched and Treg-decreased groups and found that in Treg-enriched group, highly expressed CHST11 indicated adverse prognosis (HR = 1.91, 95% CI = 1.22–3, p = 0.0039; Figure 7F). However, in Treg-decreased group, there was no significant correlation between CHST11 expression and prognosis (HR = 1.59, 95% CI = 0.86–2.54, p = 0.14; Figure 7G). These results revealed that the overexpressed CHST11 in HCC may be associated with the activation and differentiation of Tregs and thus lead to unfavorable prognosis of patients. Additionally, we also demonstrated that CHST11 expression was significantly positively correlated with immune checkpoints PD-L1 and PD-1 based on data from the TCGA and ICGC (Figure S7A-D), which could be explained by the results of KEGG (Figure S6). According to the KEGG analysis, CHST11 may participate in PD-L1 expression and PD-1 checkpoint pathway in cancer by regulating genes TICAM2, LAT, TLR2, CD4, STAT3, NFKBIE and CD3D. All of these genes were positivity correlated with CHST11 (Figure S7E) and were overexpressed in high-CHST11 expression group (Figure S7F-L). The above evidences revealed the possible relationship between CHST11 and HCC immunosuppression, providing a novel therapeutic target for HCC immunotherapy.
4 DISCUSSION
Due to an insidious onset, most of HCC patients are diagnosed at an advanced stage, leading to a clinical resectable rate of <20%.22 For patients with inoperable HCC, the 5-year survival rate is only about 8%,23 which may be partly due to tumor metastasis.24 The key prerequisite of tumor metastasis is the weakened cell adhesion ability, which is mainly regulated by adhesion-related factors on the cell surface and in the ECM. Cell surface and ECM are rich in CS, which can be covalently combined with the core protein to form CS-proteoglycan, thereby regulating cell adhesion and cell proliferation.25 CS can interact with some cell molecules (such as growth factors, adhesion molecules and cytokines) to affect disease progression.26 Increasing evidence has revealed that changing the structure and content of CS can promote tumor growth and enhance tumor invasion and metastasis.27 CS is synthesized in Golgi apparatus, and CHST11 is a key regulator for CS synthesis. Cooney et al.28 demonstrated that CS-Proteoglycan regulated by CHST11 can be used as P-selectin ligand to promote breast cancer metastasis. In addition, the carcinogenic effects of CHST11 in ovarian cancer,29 glioma,13 and leukemia30 have also been revealed. However, a study conducted by Kalathas et al.31 has presented a decreasing amount of CHST11 in colorectal cancer as the clinical stage of the cancer increases. The above evidences indicate that CHST11 plays a pro-oncogenic or tumor-suppressive role in tumor growth depending on cancer type. The role of CHST11 in HCC has not been fully understood.
This study demonstrated that CHST11 mRNA and protein both were highly expressed in HCC. The upregulated CHST11 was notably relevant to advanced TNM stage, TP53 mutation and unfavorable prognosis, suggesting that it could be served as a biomarker for HCC progression. Subsequent in vitro experiments revealed that silencing CHST11 inhibited HCC cell proliferation and metastasis. These evidences showed that CHST11 may exert a cancer-promoting role in HCC. To uncover the potential mechanism of action of CHST11 in HCC progression, we collected genes those were significantly correlated with CHST11. Functional enrichment analyses indicated that these genes not only participated in tumor growth and metastasis-related pathways, but also participated in immune-related pathways, indicating the potential immune regulatory role of CHST11 in HCC.
Despite the recent advancements in cancer immunotherapy, its therapeutic effect in HCC is unsatisfactory. One main reason is that complicated TME developed multiple ways to promote tumor cells to escape host immune response.32 To clarify the immunological correlation between CHST11 expression and TME and the potential molecular mechanism of HCC immune evasion, we further conducted an immune infiltration analysis. We first found that immune cells and stromal cells were significantly elevated in tumor samples with high CHST11 expression. Further analyses revealed that increased CHST11 expression presented high immune infiltration of Tregs. Tregs are a naturally occurring T cell subset with immunosuppressive effects that can maintain immunologic balance in the body by inducing tolerance to self-antigens.33 Yet in malignancies Tregs can secrete various inhibitory cytokines after being activated by TCR-mediated signal stimulation to suppress host antitumor immunity.34 Increasing studies have shown that immune surveillance escape mediated by Tregs promotes the progression of malignant tumors.35 The number and proportion of Treg cells are abnormally increased in peripheral blood lymphocytes and tumor-infiltrating lymphocytes in pancreatic cancer,36 breast carcinoma,37 gastric cancer,38 liver cancer39 and other malignant tumors.40 High Tregs ratio in the TME are associated with worse prognosis in majority of malignancies.41 Based on the above evidence, we speculated that CHST11 may regulate TME in HCC by promoting the infiltration of Tregs, thereby causing tumor cells to evade host immunity, further leading to tumor development and resulting in poor clinical outcomes. However, how CHST11 promotes Tregs infiltration remains unclear. Functional enrichment analyses indicted that CHST11 not only participated in biological processes related to T cell activation and differentiation, but also involved in the PD-1/PD-L1 signaling pathway. In addition, we also found that the expression of CHST11 was significantly positively correlated with PD-1 and PD-L1. Previous studies have demonstrated that the PD-1/PD-L1 pathway can induce the differentiation of Th1 cells into Tregs, promote the proliferation of Tregs, and maintain the immunosuppressive function of Tregs.42, 43 These findings suggested that CHST11 may promote Tregs activation and differentiation through the PD-1/PD-1 pathway, thereby regulating the immunosuppressive TME and promoting HCC progression.
At present, anti-PD-1/PD-L1 antibodies have made major breakthroughs in tumor immunotherapy, but their efficacy on solid tumors is not good, and novel efficacy strategies for combination therapy need to be identified. The application of anti-PD-1/PD-L1 antibodies can stimulate the proliferation and function of Tregs, thus mediating the occurrence of drug resistance in anti-PD-1/PD-L1 therapy,42 but the specific mechanism of action is not yet fully understood. Here, we found that CHST11 was both involved in Tregs infiltration and the regulation of PD-1/PD-L1 pathway, and may be a potential combined target for anti-PD-1/PD-L1 immunotherapy, providing a novel insight for improving the immunotherapy efficacy in solid tumors. However, there are limitations in this study. The immune-related analysis of this study is based on RNA-seq data and lack experimental validation. Next, we will validate the role of CHST11 in Tregs infiltration and anti-PD-1/PD-LI immunotherapy via more clinical samples and further in vitro and in vivo studies.
In conclusion, the present study revealed that CHST11 facilitated tumor proliferation and metastasis of HCC. Highly expressed CHST11 may be interrelated to immunosuppression in HCC patients, resulting in poor prognosis. These findings are potentially valuable in advancing our current understanding of CHST11 as a marker for HCC prognosis predicting and a molecular target for HCC immunotherapy.
AUTHOR CONTRIBUTION
XDD was mainly responsible for the experimental operations, data collection and manuscript writing. HRQ was mainly responsible for the data analysis and article revision. LJD, LMX, PYQ, HXL and DYW contributed to the data collection and figure plotting. CG contributed to the experiment supervision, study designation and manuscript revision.
ACKNOWLEDGEMENTS
This work was supported by the Guangxi Zhuang Autonomous Region Health Commission Self-Financed Scientific Research Project (Z20210764, Z20201147), the National Natural Science Foundation of China (NSFC82160762), the Natural Science Foundation of Guangxi, China (2022GXNSFBA035657, 2018GXNSFAA294025), the Guangxi Medical University College Student Innovation and Entrepreneurship Training Program Project (202110598312), the Guangxi Higher Education Undergraduate Teaching Reform Project (2020JGA146, 2021JGA142), the Guangxi Educational Science Planning key Projects (2021B167), the Guangxi Medical High-level Key Talents Training “139” Program (2020) and the Guangxi Medical University Training Program for Distinguished Young Scholars (2017). [Correction added on May 13, 2025 after first online publication. The grant number “NSFC82160862” has been corrected to “NSFC82160762” in this version.]
CONFLICT OF INTEREST
None.
ETHICS STATEMENT
The Ethics Committee of the First Affiliated Hospital of Guangxi Medical University approved this study.
PATIENT CONSENT STATEMENT
Not applicable.
CLINICAL TRIAL REGISTRATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.