Transcriptome-wide association study identifies multiple genes and pathways associated with pancreatic cancer
Abstract
Aim
To identify novel candidate genes for pancreatic cancer.
Methods
We performed a transcriptome-wide association study (TWAS) analysis of pancreatic cancer (PC). GWAS summary data were driven from the published studies of PC, totally involving 558 542 SNPs in 1896 individuals with pancreatic cancer and 1939 healthy controls. FUSION software was applied to the PC GWAS summary data for tissue-related TWAS analysis, including whole blood, peripheral blood, adipose, and pancreas. The functional relevance of identified genes with PC was further validated by Oncomine, STRING, and CluePedia tool.
Results
Transcriptome-wide association study analysis identified 19 genes significantly associated with PC, such as LRP5L (P value = 5.21 × 10-5), SOX4 (P value = 3.2 × 10-4), and EGLN3 (P value = 6.2 × 10-3). KEGG pathway enrichment analysis detected several PC-associated pathways, such as One carbon pool by folate (P value = 1.60 × 10-16), Cell cycle (P value = 1.27 × 10-7), TGF-beta signaling pathway (P value = 4.64 × 10-6). Further comparing the 19 genes with previously identified overexpressed genes in PC patients found one overlapped gene SOX4.
Conclusion
We identified some novel candidate genes and pathways associated with PC. Our results provide novel clues for the genetic mechanism studies of pancreatic cancer.
1 INTRODUCTION
Pancreatic cancer(PC) is a deadly disease, which leads to an estimated 227 000 deaths per year worldwide.1 The term pancreatic cancer contains both exocrine and endocrine tumors of pancreas. More than 90% of PCs originate from ductal epithelium.2 Risk factors for this malignant disease include smoking, advanced age, family history of chronic pancreatitis, male sex, diabetes, obesity, non-O blood group, occupational exposures, diets high in meat and low in vegetables, and probable Helicobacter pylori infection and periodontosis.1
Although the cause of PC is multifactorial, the most important risk factors are smoking and family history.Nearly 20% of pancreatic tumors are caused by smoking, and cancers from smokers suffer more genetic mutations than those from nonsmokers.1 Genetic factors play important roles in the development of pancreatic cancer. Multiple genetic studies have been implemented and identified some susceptibility genes for PC, such as KRAS, BRCA1, BRCA2, PALB2, FANCC, FANCG, ATM, CDKN2A, PRSS1, SPINK1, TERT, NR5A2, ZNRF3, and SMC2.3 However, the genetic risk of PC explained by the reported candidate loci was limited, indicating the existence of undiscovered susceptibility loci for PCs.
Genome-wide association studies (GWAS) are an efficient tool for genetic mechanism studies of complex diseases. Expression quantitative trait loci (eQTLs) are a group of important regulatory loci, which can regulate gene expression levels.4 In recent years, integrative analysis of GWAS data and eQTLs annotation information is rapidly becoming a standard approach for explore the genetic basis of disease susceptibility.5 Nowadays, transcriptome-wide association study (TWAS) analysis was proposed to utilize extensive published GWAS data. TWAS analysis adopts pre-computed gene expression weights together with disease GWAS summary statistics to estimate the association of each gene to diseases.6, 7 TWAS showed a high efficiency for identifying novel causal genes of complex diseases.8-11
In this study, we conducted a tissue-related TWAS for PC, considering whole blood, peripheral blood, adipose, and pancreas. TWAS was first applied to a large-scale GWAS data to detect novel susceptibility genes associated with PC. The functional relevance of identified genes with PC was further validated by Oncomine, STRING, and CluePedia tool.
2 METHODS
2.1 GWAS summary datasets of PC
A large-scale GWAS summary data of pancreatic cancer were used in this study.7 Briefly, this GWAS comprised of 1896 individuals with pancreatic cancer and 1939 controls from 12 prospective cohorts and a hospital-based case-control study. Samples were excluded based on low completion rates (<98%) and unexpected inter- or intra-study duplicates. Commercial platform Illumina Hap500 Infinium genotyping assay was used for genome-wide SNP genotyping. GLU, Genotyping Library and Utilities, was used for data analysis and management. The association between single SNPs and pancreatic cancer was tested by Logistic regression. SNPs were excluded based on low call rates (<90%). Detailed information of cohorts, genotyping, and quality control approaches can be found in the published studies.7
2.2 Gene expression datasets
The over-expression genes in PC patients were driven from the Oncomine database.12 Oncomine (https://www.oncomine.org) is a cancer microarray database and web-based data-mining platform for facilitating discovery from genome-wide expression analyses. Differential gene expression was identified by comparing major types of cancer to respective normal tissues.12, 13
2.3 TWAS of pancreatic cancer
FUSION software was applied to the PC GWAS summary data for tissue-related TWAS analysis, including whole blood, peripheral blood, adipose, and pancreas. TWAS analysis uses pre-computed gene expression weights together with disease GWAS summary statistics to calculate the association of every gene to disease.14 The genetic values of expression were computed one probe set at a time using SNP genotyping data located 500 kb on either sides of the gene boundary. For this study, the gene expression weights of whole blood, peripheral blood, adipose, and pancreas were driven from the FUSION website (https://gusevlab.org/projects/fusion/).14 The genes with significant and suggestive association signals were identified at P value <3.73 × 10-6 after strict Bonferroni correcting and P value <0.05, respectively.
2.4 PPI network and pathway enrichment analysis
The functional relevance of identified genes with PC was further validated byOncomine, STRING, and CluePedia tool. STRING (Search Tool for the Retrieval of Interacting Genes) (https://string-db.org/cgi/input.pl) is an online tool designed to evaluate the protein-protein interaction (PPI) network.15, 16 The CluePedia, a plugin of Cytoscape software, is a tool for searching potential genes associated with the certain signaling pathway by calculating linear and nonlinear statistical dependencies from experimental data.17, 18 The PPI (protein-protein interaction) network of significant genes identified by TWAS was constructed by STRING. We also analyzed the signaling pathways of these significant genes by STRING, then verified, and visualized them by CluePedia. The candidate pathways were identified at P value <0.05.
3 RESULTS
3.1 TWAS analysis results
A total of 13 412 genes were analyzed by TWAS in this study. After strict Bonferroni correcting, TWAS identified 19 genes with suggestive association signals (P value <0.05) (Table 1), such as LRP5L (P value = 5.21 × 10-5), SOX4 (P value = 3.2 × 10-4), EGLN3 (P value = 6.2 × 10-3), TXNDC15 (P value = 6.5 × 10-3), PPP2R2A (P value = 9.7 × 10-3), and E2F3(P value = 4.4 × 10-2).
| Gene | Chromosome | GWAS SNP ID | EQTL ID | TWAS P value | Comparative tissue |
|---|---|---|---|---|---|
| KCNK5 | 6 | rs2162840 | rs228815 | 4.67E-02 | Adipose |
| RP11-65J3.1 | 9 | rs10819503 | rs17457362 | 4.67E-02 | Adipose |
| PPP2R2A | 8 | rs6557943 | rs10495 | 9.70E-03 | Adipose |
| PPFIBP2 | 11 | rs1552462 | rs10839816 | 3.47E-02 | Adipose |
| GEMIN4 | 17 | rs4985584 | rs3744743 | 1.71E-02 | Adipose |
| NIPA2 | 15 | rs17524529 | rs7168115 | 3.82E-02 | Peripheral blood |
| RNASEH2B | 13 | rs7985384 | rs9568490 | 3.98E-02 | Peripheral blood |
| FARS2 | 6 | rs6920599 | rs2224391 | 3.11E-02 | Whole blood |
| E2F3 | 6 | rs4712465 | rs874447 | 4.36E-02 | Whole blood |
| SOX4 | 6 | rs12530233 | rs12530233 | 3.22E-04 | Whole blood |
| MTHFD1L | 6 | rs13201018 | rs13201018 | 1.09E-02 | Whole blood |
| F2R | 5 | rs4438855 | rs153311 | 3.95E-02 | Whole blood |
| TXNDC15 | 5 | rs6889478 | rs12656013 | 6.46E-03 | Whole blood |
| NDUFA3 | 19 | rs254260 | rs254262 | 1.50E-02 | Whole blood |
| CRISPLD2 | 16 | rs1566465 | rs8057431 | 3.21E-02 | Whole blood |
| CHSY1 | 15 | rs4965799 | rs2012387 | 1.78E-02 | Whole blood |
| EGLN3 | 14 | rs1680699 | rs1769603 | 6.24E-03 | Whole blood |
| IQSEC3 | 12 | rs12582189 | rs2291926 | 1.92E-02 | Whole blood |
| LRP5L | 22 | rs5761251 | rs13057190 | 5.21E-05 | Whole blood |
3.2 PPI network and pathway enrichment analysis
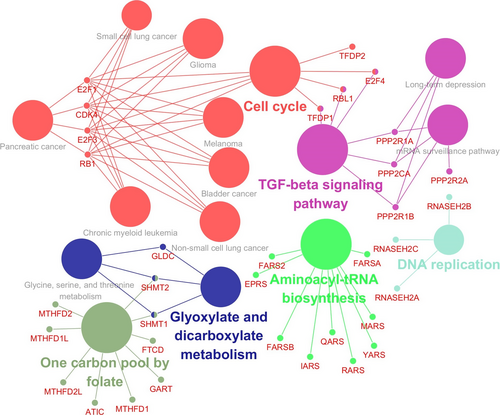
The PPI network and signaling pathways of the 19 PC-associated genes were generated by STRING (Table 2). The top 10 candidate pathways identified by the STRING tool were one carbon pool by folate (P value = 1.60 × 10-16), aminoacyl-tRNA biosynthesis (P value = 4.64 × 10-13), metabolic pathways (P value = 2.07 × 10-7), cell cycle (P value = 1.27 × 10-7), TGF-beta signaling pathway (P value = 4.64 × 10-6), bladder cancer (P value = 1.71 × 10-5), purine metabolism(P value = 2.75 × 10-5),non-small cell lung cancer (P value = 5.95 × 10-5),pancreatic cancer (P value = 7.72 × 10-5),glioma(P value = 7.72 × 10-5). The results showed that relevant genes of PC were CDK4, E2F1, E2F3, and RB1 (Table 2). Then, we verified and visualized the linkage of genes with PC by CluePedia (Figure 1). The genes enriched to PC were coincident with the results from STRING.
| Pathway description | Count | STRING P value | Gene ID | CluePedia P value |
|---|---|---|---|---|
| One carbon pool by folate | 9 | 1.60E-16 |
ATIC,FTCD,GART,MTHFD1,MTHFD1L,MTHFD2, MTHFD2L,SHMT1,SHMT2 |
19.0E-16 |
| aminoacyl-tRNA biosynthesis | 9 | 4.64E-13 |
EPRS,FARS2,FARSA,FARSB,IARS,MARS,QARS, RARS,YARS |
34.0E-11 |
| Metabolic pathways | 18 | 1.27E-07 |
ADSL,ATIC,CHSY1,EPRS,FTCD,GART,GLDC, MTHFD1,MTHFD1L,MTHFD2,MTHFD2L, NDUFA3,PAICS,PFAS,PPAT,QARS,SHMT1,SHMT2 |
0.0026 |
| Cell cycle | 8 | 1.27E-07 | CDK4,E2F1,E2F3,E2F4,RB1,RBL1,TFDP1,TFDP2 | 18.0E-7 |
| TGF-beta signaling pathway | 6 | 4.64E-06 | E2F4,PPP2CA,PPP2R1A,PPP2R1B,RBL1,TFDP1 | 0.00004 |
| Bladder cancer | 4 | 0.000171 | CDK4,E2F1,E2F3,RB1 | 0.00052 |
| Purine metabolism | 6 | 0.000275 | ADSL,ATIC,GART,PAICS,PFAS,PPAT | 0.0032 |
| Non-small cell lung cancer | 4 | 0.000594 | CDK4,E2F1,E2F3,RB1 | 0.0014 |
| Pancreatic cancer | 4 | 0.000772 | CDK4,E2F1,E2F3,RB1 | 0.0024 |
| Glioma | 4 | 0.000772 | CDK4,E2F1,E2F3,RB1 | 0.0024 |
| Tight junction | 5 | 0.000783 | CDK4,PPP2CA,PPP2R1A,PPP2R1B,PPP2R2A | 0.0035 |
| Melanoma | 4 | 0.000968 | CDK4,E2F1,E2F3,RB1 | 0.0024 |
| Chronic myeloid leukemia | 4 | 0.000968 | CDK4,E2F1,E2F3,RB1 | 0.0022 |
| Small cell lung cancer | 4 | 0.00194 | CDK4,E2F1,E2F3,RB1 | 0.0034 |
| mRNA surveillance pathway | 4 | 0.00199 | PPP2CA,PPP2R1A,PPP2R1B,PPP2R2A | 0.0023 |
3.3 Identification of overexpressed genes in SCLC
We identified overexpressed and down-expressed genes in PC by comparing the PC samples to normal tissues in Oncomine online database. The top 1% genes overexpressed and down-expressed in 78 PC samples were shown in Appendix S1. After comparing the genes identified by TWAS with the overexpressed and down-expressed genes detected by Oncomine, we found one overlapped gene SOX4, which was overexpressed in PC patients. Furthermore, the significant SNP rs12530233 of SOX4 gene was also an eQTL, suggesting the functional importance of rs12530233 in the development of PC.
4 DISCUSSION
Pancreatic cancer is the main cause of cancer death worldwide. The cause of pancreatic cancer is complex and multifactorial. Smoking, advanced age, and family history of chronic pancreatitis are main risk factor for PC.1 Most patients with pancreatic cancer remain asymptomatic until the tumor metastasizes to other tissues and organs.19 As there is no standard program for screening patients at early-stage of PC, it is necessary to find more effective susceptibility gene for PC prevention.
Consistent with the result of TWAS, previous studies have reported 4 of the 19 genes (PPP2R2A, E2F3, KCNK5, and SOX4) play important roles in the development of PC,20-23 and 2 of the 19 genes (CHSY1 and EGLN3) have been proved associated with PC via bioinformatics methods.25, 26 SOX4 is significantly associated with PC and overexpressed in PC patients. Furthermore, it is interesting that the significant SNP rs12530233 of SOX4 gene is an eQTL, suggesting the importance of rs12530233 in the dysfunction of SOX4 expression regulation during the development of PC. Previous research has verified that SOX4 is expressed in the early processes of PC tumorigenesis and suggested that SOX4 might function as a master transcription factor in PC formation.24 Further studies, such as fine mapping and RNA sequencing of SOX4, are needed to confirm our finding and clarify the potential mechanism of SOX4 involved in the development of PC.
Another susceptibility gene E2F3, is a transcription factor family, plays an important role in cellular proliferation, apoptosis, and differentiation.27 MiR-210 is induced by hypoxia and expressed in the development of PC. Chen et al28 have suggested that E2F3 may be potential miR-210 targets in PC. A systems-level analysis of the scale-free GMCs network taken by Rajamani et al29 identified that E2F3 is associated with PC progression.
It has been known that PPP2R2A encodes an alpha isoform from the regulatory subunit B55 subfamily (B55α) and can selectively control Akt phosphorylation.30 Intriguingly, Hein et al21 have reported that PPP2R2A promotes PC development by maintaining hyperactivity of multiple oncogenic signaling pathways, including ERK, Wnt as well as AKT. In addition, Shen et al22 have found PPP2R2A was significantly higher expressed in SH-PAN cells than DT-PCa cells and proved that decreased expression of PPP2R2A inhibited the development and progression of PC.
KCNK5 (also known as TASK-2 or K2P5.1) has been shown to be the volume sensitive K (+) channel in cells.31 KCNK5 is expressed in the kidney, liver, stomach, small intestine, colon, and pancreatic acinus. An electrophysiological study indicated that KCNK5 was expressed in human pancreatic ductal adenocarcinoma cell line, and the pH-sensitive K2P subunits coded by KCNK5 were shown to be expressed in pancreatic.20
Transcriptome-wide association study analysis identified several novel candidate genes for PC, such as RP11-65J3.1, PPFIBP2, GEMIN4, NIPA2,RNASEH2B, FARS2, MTHFD1L, F2R, TXNDC15, NDUFA3, CRISPLD2, IQSEC3, and LRP5L. As is known to all, few efforts have been paid to investigate the potential roles of them in the formation of PC. Further biological experimental studies are required to confirm our finding and clarify the potential roles of novel candidate genes in the development of PC.
Pathway enrichment analysis detected several candidate pathways for PC, some of which has been reported to be implicated in the development of PC directly or indirectly. For instance, it has been demonstrated that TGF-beta signaling pathway in pancreatic cancer can be utilized in targeted therapy clinical trials.32 Another interesting pathway is one carbon pool by folate, which is associated with influenced PC incidence.33 Researchers have also proved that aminoacyl-tRNA biosynthesis, metabolic pathways, cell cycle, tight junction, purine metabolism, and mRNA surveillance pathway are existed in PC cells and related to occurrence and development of PC.33-38 The rest of these signaling pathways are solid tumor directly. Like bladder cancer, lung cancer, glioma, melanoma, chronic myeloid leukemia as well as PC.
In summary, we conducted a tissue-related TWAS analysis and identified some novel candidate genes and pathways associated with PC. Our results provide novel clues for clarifying the genetic mechanism of PC. Further biological studies are warranted to confirm our findings and reveal the potential mechanism of identified genes and pathways involved in the development of PC.
ACKNOWLEDGMENT
This work was supported by grants from National Natural Science Foundation of China 81672921, Innovation Capacity Support Plan of Shaanxi Province 2018TD-002.
CONFLICT OF INTEREST
The authors declare no conflict of interests.




