m6A-mediated regulation of PBX1-GCH1 axis promotes gastric cancer proliferation and metastasis by elevating tetrahydrobiopterin levels
Abstract
Background
Methyltransferase 3 (METTL3)-mediated N6-methyladenosine (m6A) RNA modification has been demonstrated to be a potential factor in promoting gastric cancer (GC). METTL3 regulates a series of signaling pathways by modifying various mRNAs. This study aimed to identify novel METTL3-mediated signaling pathways and explored possible targets for use in the clinical setting of gastric cancer.
Methods
To investigate the proliferation and metastatic capacity of GC cell lines with METTL3 knockdown, a xenograft, lung metastasis, and popliteal lymph node metastasis model was used. The m6A-modified RNA immunoprecipitation (Me-RIP) sequence was utilized to explore the target mRNAs of METTL3. Cell counting kit 8 and transwell assays were performed to investigate the promoting function of pre-B cell leukemia homeobox 1 (PBX1) and GTP cyclohydrolase 1 (GCH1). Western blotting and chromatin immunoprecipitation were employed to confirm the involvement of the METTL3-PBX1-GCH1 axis. ELISA and liquid chromatography-mass spectrometry were used to explore the biological function of tetrahydrobiopterin (BH4).
Results
Knockdown of METTL3 suppressed xenograft tumor growth and lung/lymph node metastasis in vivo. Mechanistically, we found that METTL3 combined with and stabilized PBX1 mRNAs. Chromatin immunoprecipitation (ChIP) and further experiments suggested that PBX1 acted as a transcription factor inducing GCH1 expression. Moreover, the METTL3-PBX1-GCH1 axis increased BH4 levels in GC cells, thereby promoting tumor progression.
Conclusions
This study suggested that METTL3 enzymes promote tumor growth and lung/lymph node metastasis via METTL3-PBX1-GCH1 axis increasing BH4 levels in GC.
List of Abbreviations
-
- 3′ UTR
-
- 3′ untranslated region
-
- 5′ UTR
-
- 5′ untranslated region
-
- ALKBH5
-
- alkB homolog 5
-
- BH4
-
- Tetrahydrobiopterin
-
- CCK-8
-
- Cell count kit 8
-
- ChIP
-
- Chromatin immunoprecipitation
-
- DMSO
-
- Methyl sulfoxide
-
- FBS
-
- fetal bovine serum
-
- GAPDH
-
- glyceraldehyde-3-phosphate dehydrogenase
-
- GC
-
- gastric cancer
-
- GCH1
-
- GTP cyclohydrolase 1
-
- GTEx
-
- Genotype-Tissue Expression
-
- H&E
-
- hematoxylin and eosin
-
- IF
-
- Immunofluorescence
-
- IHC
-
- immunohistochemistry
-
- LC-MS
-
- Liquid Chromatography with tandem mass spectrometry
-
- m6A
-
- N6-methyladenosine
-
- MB
-
- Methylene blue
-
- Me-RIP
-
- m6A-modified RNA immunoprecipitation
-
- METTL14
-
- methyltransferase like 14
-
- METTL3
-
- methyltransferase 3
-
- one-way ANOVA
-
- one-way analysis of variance
-
- OS
-
- Overall survival
-
- PBS
-
- Phosphate Buffered Saline
-
- PBX1
-
- Pre-B-cell leukemia homeobox 1
-
- PFS
-
- Progression free survival
-
- RIP
-
- RNA immunoprecipitation
-
- seq
-
- sequence
-
- shM3
-
- Knockdown of METTL3
-
- shNC
-
- Negative control of knockdown
-
- siRNA
-
- small interfering RNA
-
- SPF
-
- Specific Pathogen Free
-
- SPRi3
-
- SPR inhibitor 3
-
- TCGA
-
- The Cancer Genome Atlas Program
-
- ZNF668
-
- zinc finger protein 668
-
- β-actin
-
- actin beta
1 INTRODUCTION
Gastric cancer (GC) has become the sixth most common cancer worldwide, with an estimated 1,089,103 cases and about 700,000 deaths by 2020 [1, 2]. GC development is a complex process, including crosstalk between genetic mutations, epigenetic modifications, metabolic reprogramming, and microbiome [3]. In clinical practice, numerous therapy options for GC treatment have been adopted, including surgery, chemotherapy, anti-angiogenic therapy, and immunotherapy [4, 5]. However, the 5-year survival rate of patients with GC is still lower than 50% [6]. Thus, it is crucial to explore the mechanism of GC development and identify novel targets for GC treatment.
Pre-B-cell leukemia homeobox 1 (PBX1) interacts with PREP1 or MEIS1 to form a dimer. The PREP1/MEIS1-PBX1 dimer translocates into the nucleus depending on the nuclear localization signal (NLS) provided by PBX1 and acts as a transcription factor (TF) [7]. In contrast, PBX1 is a cofactor of HOX proteins and regulates axial patterning [8]. The gene was first reported as an oncogene in acute lymphoblastic leukemia, which gained the carcinogenic effect of the t(1;19) juxtaposes, resulting in E2A and PBX1 fusion [9]. Recent studies also found that a dominant-negative Pbx1 mutant, Pbx1NT, is required for the oncogenic activity of HOXB7 in melanoma and breast adenocarcinoma [10]. In addition, PBX1 functions as an activated TF that upregulates lipid metabolism genes in estrogen receptor-negative breast cancer [11]. It has been reported that the PBX1 protein level is elevated in the gastric mucosa of GC patients with Helicobacter pylori (H. pylori) infection [12]. However, the mechanism underlying the change in PBX1 levels remains unknown, and the biological significance of upregulated PBX1 expression is yet to be explored.
N6-methyladenosine (m6A) has been proven to be the most abundant modification of RNA in eukaryotic cells [13]. The m6A motifs regulate the stability, splicing, location, and translation of the target RNA. This epigenetic modification is reversible, installed by ‘writer’, proteins and eliminated by ‘eraser’ proteins, such as ALKBH5 and FTO. METTL3, METTL14 and WTAP form the m6A methyltransferase complex responsible for most m6A modifications. In recent years, METTL3 has been reported to have a notable effect in different typical cancers, including prostate [14], esophageal [15], and lung cancer [16]. In gastric adenocarcinoma, Wang et al. [17] reported that METTL3 stabilized HDFG mRNAs to affect the aerobic glycolysis of GC cells. Other studies have shown that ARHGAP5, α-smooth muscle actin and ZMYM1 are also target genes of METTL3, affecting the biological behaviors of a series of GCs [18-20]. These results indicate that the biological effects of MEETL3 are diverse, depending on its downstream genes.
Tetrahydrobiopterin (BH4), involved in endogenous synthesis of GTP by GTP cyclohydrolase-1 (GCH1), is also involved in the production of monoamine neurotransmitters, in the generation of nitric oxide, and in the process of pain by acting as an enzyme cofactor [21, 22]. BH4 production increases in T cells, which is vital for their proliferative ability and anti-tumor activity [23]. BH4 induces lipid remodeling in fibrosarcoma and protects cells from ferroptosis [24]. Specific inhibitors of BH4 biosynthesis, such as DAHP, SPRi3, and QM385 [23], have been designed, and their efficiency has been widely proven [24]. Thus, understanding the role of BH4 in GC has potential value in pharmacology.
In the present study, we revealed an innovative m6A-mediated PBX1-GCH1 pathway that regulates the biosynthesis of BH4. These findings suggest that METTL3 and BH4 may be new targets for GC therapy.
2 MATERIALS AND METHODS
2.1 Patients and tissue specimens
Tumor and paired para-carcinoma tissues were collected from 100 patients with GC, diagnosed at the First Affiliated Hospital of Sun Yat-sen University (Guangzhou, Guangdong, China) between January 2011 and December 2016. All patients were diagnosed with GC preoperatively and underwent radical excision. Patients who died within three months after surgery or had more than one primary tumor been excluded. All tissue specimens were separated and frozen at -80℃ or formalin-fixed and paraffin-embedded. Experienced pathologists from the First Affiliated Hospital of Sun Yat-sen University reviewed the tissue sections. Ethical approval for research on human subjects was obtained from the Institutional Review Board of the First Affiliated Hospital of Sun Yat-sen University. Each participant signed an informed consent form before participating in the study.
2.2 Cell lines, cell culture, and transfection
Human gastric cancer cell lines (AGS, MKN1, SGC7901, and MGC803) were purchased from the Chinese Academy of Sciences Shanghai Branch Cell Bank (Shanghai, China). AGS cells were cultured in DMEM/F12 (Gibco, ThermoFisher, Waltham, MA, USA) and MKN1, SGC7901, and MGC803 cells were cultured in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum (FBS; Biological Industries, Israel) and 1% antibiotics (penicillin/streptomycin, Gibco) at 37°C with 5% CO2. The cell lines were tested for potential mycoplasma contamination by PCR and confirmed mycoplasma-negative.
Drug treatments, such as STM2457 (MedChemExpress, New Jersey, USA), were performed as previously described [25]. Briefly, the cells were grown on 6 well culture plates with 10% FBS medium 24 h before STM2457 treatment. Then, STM2457 was added to the medium at a concentration of 10 μmol/L.
According to the manufacturer's instructions, siRNA transfection was performed using Lipofectamine RNAi Max (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA). Briefly, siRNAs were diluted in the 100 μL Opti-MEM (Gibco) at 5 nmol/L and mixed with 5 μL Lipofectamine RNAi Max. After incubation at room temperature for 20 min, the mixture was added to the cell culture medium, and the cells were collected after 48 h. The sequences of specific siRNAs are listed in Supplementary Table S1. Overexpression plasmids were constructed using pEZ-Lv201 (iGene Biotechnology, Guangzhou, Guangdong, China), and the knockdown plasmids were constructed using LVRU6GP (iGene Biotechnology). All plasmids were constructed by iGene Biotechnology Co., Ltd. Transfection of plasmids was performed using Lipofectamine 3000 (Invitrogen). Briefly, 2 μg plasmids and 5 μL Lipofectamine 3000 were diluted in 150 μL Opti-MEM and 5 μL Lipofectamine 3000 were diluted in 150 μL Opti-MEM. Then, the plasmids and Lipofectamine 3000 solution were mixed and incubated at room temperature for 15 min. Finally, the mixture was added onto 6 well plates, and cells were collected after 48-72 hours for further studies.
2.3 Animals and animal models
BALB/c male nude mice (5-6 weeks old) were purchased from Specific Pathogen Free (Beijing) Biotechnology Co., Ltd. (Beijing, China) and maintained in specific pathogen-free facilities. The Institutional Ethical Board approved animal studies of the First Affiliated Hospital of Sun Yat-sen University.
For tumor xenograft models, MGC803 cells (5×106) with METTL3 knockdown and negative control were subcutaneously injected into the right axilla of nude mice (n = 6 per group). Tumor volume was monitored every other day (volume = length × width2 × 1/2). When the largest tumor diameter reached 1 cm, the mice were sacrificed via a cervical dislocation manner. The tumors were weighed, imaged, and fixed in 4% paraformaldehyde or frozen for further analyses. To investigate the function of SPRi3, xenograft models were first established using MGC803 cells overexpressing METTL3 and a blank control. After one week, SPRi3 (300 mg/kg in 10 μL DMSO; Solarbio, Beijing, China) was injected into the tumors, and an equal volume of DMSO was injected as a negative control. The injection was performed weekly until the mice were sacrificed after 8 weeks. The tumors were resected, photographed, and fixed in 4% paraformaldehyde for further analysis.
For the lung metastasis model, MGC803 cells (luciferase labeled, 1×106) with METTL3 knockdown and negative control were injected intravenously into the tail vein of nude mice. Six weeks after injection, fluorescein sodium salt (Solarbio) was injected into the tail vein of the mice. The bioluminescence images were captured using an IVIS Lumina II (Xenogen Corp., Alameda, California, USA). After 8 weeks, the mice were sacrificed by cervical dislocation, and the lungs were resected, photographed, and fixed in 4% paraformaldehyde for further analyses.
For the popliteal lymph node metastasis model, MGC803 cells (Luciferase labeled, 1×106 in 20 μL PBS) were injected into the left footpad of mice (twice in three days). Six weeks after injection, fluorescein sodium salt (Solarbio) was injected into the tail vein of the mice. The bioluminescence images were captured using an IVIS Lumina II (Xenogen Corp.). After 8 weeks, the mice were sacrificed by cervical dislocation. The left legs' footpad and popliteal lymph nodes were resected, photographed, and fixed in 4% paraformaldehyde for further analysis.
2.4 RNA extraction and real-time PCR (qPCR) analysis
According to the manufacturer's instructions, total RNA was extracted from cells and tissues using RNAiso (Takara, Kusatsu, Japan). Briefly, the cells in 6 well plates were lysed in 500 μL RNAiso on ice and then mixed with 200 μL chloroform(Baishi, Guangzhou, Guangdong, China). After incubating for 10 min on ice, the mixture was centrifuged at 12000 rpm for 15 min, and the chloroform was collected. Next, 500 μL isopropanol (Baishi) was added to the mixture and incubated for 10 minutes. After centrifugation at 12000 rpm for 10 min, the RNAs were placed at the bottom of the tubes. Then, the RNA precipitate was washed with 75% ethyl alcohol (Baishi) and diluted in diethyl pyrocarbonate (DEPC) water (Takara).
As instructed, quantifications of specific RNA were used SYBR® Green I (Accurate Biotechnology, Changsha, Hunan, China). Briefly, 5 μL SYBR Green, 3 μL DEPC water, 1 μl primers and 1 μl cDNA were prepared for each sample, and the qPCR protocol was run using a LightCycler® 480 II (Roche, Basel, Switzerland). The fold change was calculated using the relative quantification method (2−ΔΔCt). The qPCR primers used are listed in Supplementary Table S2.
2.5 Cell proliferation, colony formation, migration, and invasion assays
The cells were seeded in 96-well plates at 1,000 cells per well for the cell proliferation assays. Cell proliferation was evaluated using 10% CCK-8 (SolarBio) diluted in standard culture media for 2 h. Proliferation rates were determined at 0, 24, 48, 72, and 96 h after seeding, and quantification was performed on a microtiter plate reader (ALLSHENG, Hangzhou, Zhejiang, China) measured at UV wavelength (λ) = 450 nm.
For colony formation assay, cells were seeded in 6-well plates at a density of 500 cells/well. The cells were then cultured in complete media for 4-6 weeks until the size of a single colon reached 2-5 mm. Then the colonies were fixed by 4% paraformaldehyde and stained by 1% leucocrystal violet for 5 minutes.
For the cell migration and invasion transwell assays, 50,000 cells in 400 μL media with 10% fetal bovine serum were plated on the top chambers of Transwell Clear Polyester Membrane Inserts (for the migration assay, Corning Costar, New York, USA) and BioCoat Matrigel Invasion Chambers (for the invasion assay, Corning Costar), while culture media with 10% FBS was applied to the bottom. After 16-72 hours, migrated or invaded cells were stained with crystal violet and counted under a microscope (Olympus, Tokyo, Japan).
2.6 m6A dot blot assay
The dot blot assay was performed according to an online protocol (https://en.bio-protocol.org/e2095). Briefly, RNA was isolated from cells as previously described and denatured at 95°C for 2 min. Then, 2000 ng of mRNA was spotted onto a Hybond-N+ membrane (Cytiva, Shanghai, China) and crosslinked by UV (4000 × 100 μJ/cm2, twice; Spectronics, USA). The membrane was then washed and incubated in blocking buffer (5% skim milk, EpiZyme, Guangzhou, Guangdong, China) and then gently shaken overnight with an anti-m6A antibody (synaptic system, 202003; Goettingen, Germany) at 4°C. The membrane was then washed, incubated with an anti-mouse antibody (EpiZyme), and washed again. Finally, the membrane was exposed to Hyperfilm ECL (Thermo Fisher, Waltham, MA, USA), and images were acquired using a ChemiDoc™ imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Methylene blue (MB Solarbio) was used to interact with mRNA and as a loading control.
2.7 m6A-modified RNA immunoprecipitation (Me-RIP) and RNA immunoprecipitation (RIP)
Cells in 150 mm dishes were crosslinked by UV (4000 × 100 μJ/cm2, twice; Spectronics) and collected in PBS. The RNA-m6A complexes were isolated using 0.5% NP-40 (Sigma-Aldrich, Merck Millipore, Darmstadt, Germany). After pelleting DNA via centrifugation, the suspension was added to 5 μg anti-m6A (for Me-RIP, synaptic system), anti-METTL3 (for RIP, Proteintech), or anti-IgG (EpiZyme) and incubated overnight at 4°C. Then, 40 μL protein A/G magnetic beads (Invitrogen, Thermo Fisher Scientific, Inc.) were added to the mixture for 1 h, and the beads were collected and washed twice with wash buffer (0.1% SDS and 0.5% NP-40 in PBS, Gibco). The RNA-m6A complex was de-crosslinked by heating at 45°C for 1 h. Finally, RNAiso (Takara) was added to the beads for RNA isolation. qPCR measured the quantification of m6A-enriched PBX1. The enrichment percentage was calculated using the relative quantification method: 2^ (CTIP-CTinput).
2.8 m6A-modified RNA immunoprecipitation-sequencing (MeRIP-seq)
For MeRIP-seq, the RNAs were purified by Spin Column RNA Cleanup & Concentration Kit (Sangon Biotech, Shanghai, China). Total RNA was greater than 120 μg, and the integrity and quantity of each RNA sample were assessed using agarose gel electrophoresis and a NanoDropTM instrument (ThermoFisher). According to the manufacturer's protocol, intact mRNA was first isolated from total RNA samples using an Arraystar Seq-StarTM poly(A) mRNA Isolation Kit (Aksomics, Shanghai, China). The isolated mRNA was chemically fragmented into 100-nucleotide-long fragments by incubation in fragmentation buffer (10 mmol/L Zn2+ and 10 mmol/L Tris-HCl, pH 7.0), and the size of the fragmented mRNA was confirmed using agarose gel electrophoresis. Then, m6A-methylated mRNAs were immunoprecipitated with an anti-N6-methyladenosine antibody (synaptic system), as previously described, and an aliquot of the fragmented mRNAs was collected. The major procedures included immunoprecipitation, washing, and elution. The eluted m6A mRNA fragments were then concentrated for RNA-seq library construction. A KAPA Stranded mRNA-seq Kit (F. Hoffmann-La Roche Ltd, Grenzacherstrasse, Basel, Switzerland) was used to create RNA-seq libraries for m6A antibody-enriched and input mRNAs (Illumina, San Diego, CA, USA). The prepared libraries were diluted to a final concentration of 8 pmol/L, and clusters were generated on an Illumina cBot using a HiSeq 3000/4000 PE Cluster Kit (#PE-410-1001, Illumina), followed by sequencing on an Illumina HiSeq 4000. Finally, all reads were mapped to the human genome 19 using BWA tools and annotated using miRBase version 21 (https://www.mirbase.org) and miRDeep2 (https://www.mdc-berlin.de/8551903/en/). The data were analyzed using R software (Free Software Foundation, Boston, MA, USA).
2.9 Chromatin immunoprecipitation (ChIP)
The ChIP assay was performed using a ChIP assay kit (SimpleChIP® Plus Sonication Chromatin IP Kit (#56383, Cell Signaling Technology, MA, USA). Briefly, 5×106 GC cells were fixed with 1% formaldehyde (Baishi), quenched with glycine (#G8200, Solarbio) at room temperature, following which they were collected, washed, and resuspended in lysis buffer (Cell Signaling Technology). The sonicated chromatin solution was immunoprecipitated with anti-IgG (DIA-AN, Q6004; Wuhan, Hubei, China) and anti-PBX1 (Abnova, H00005087-M01; Tsukiji, Tokyo, Japan). Immunoprecipitated DNA was purified by column collection and analyzed by qPCR. The enrichment percentage was calculated using the relative quantification method: 2CTChIP-CTinput.
2.10 The bioinformatics analysis of online database
We downloaded the clinical data from the database of The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/) and Genotype-Tissue Expression (GTEx) (https://gtexportal.org/home/). The differentially expressed genes were calculated in R version 4.1.2 with EdgeR package [26]. CHIPBbase v2.0 [27] and JASPAR database [28] were used to identify potential downstream genes of transcription factors. For CHIPBase analysis, target transcription factors were imported and the binding region was chosen as “up/down 1000bp”. All the genes output from the database were downloaded for further analysis. For JASPR database, target transcription factors were selected and the sequence of 1000bp upstream of potential downstream genes were imported in “Scan” box. Relative profile score threshold was 80%.
2.11 Untargeted sequence of metabolism
Novogene Co., Ltd. (Beijing, China) performed the untargeted metabolomics sequencing. Briefly, the samples were placed in EP tubes and resuspended with prechilled 80% methanol (Baishi) and 0.1% formic acid (Rhawn, Shanghai, China) by vortexing. The samples were then melted on ice and washed for 30 s. After sonification for 6 min, the samples were centrifuged at 5,000 rpm at 4°C for 1 min. The supernatant was freeze-dried and dissolved in 10% methanol. Finally, the solution was injected into the Liquid Chromatography with tandem mass spectrometry (LC-MS-MS) system analysis (SCIEX Headquarters, Framingham, MA, USA).
2.12 Enzyme-linked immunosorbent assay (ELISA)
The NO levels in cells were detected by colorimetric ELISA-like assay via the Nitric Oxide (NO) assay kit (BioAssay Systems; Hayward, CA, USA) according to the manufacturer's instructions. Briefly, the cell lysis was added in 96-well plates and incubated with antibody from the kit. After washing the antibody away, reaction reagents were added and the absorbancy at λ = 405 mm was read.
2.13 Statistical Analysis
Overall survival (OS) is defined as the interval between the first radical resection to the date of death from any cause or to the date of the last follow-up visit. And the Progression-Free-Survival (PFS) is defined as the interval between the first radical resection to the first disease progression or death. Kaplan-Meier analysis was used to analyze the survival fraction.
All statistical values were calculated using Statistical Product and Service Solutions (SPSS) 22.0 (Chicago, IL, USA). Experimental studies were analyzed by independent sample t-test to compare two groups and by one-way ANOVA followed by post hoc Tukey's test to compare multiple groups. Kaplan-Meier analysis and log-rank tests were used to evaluate differences in patient survival. The Spearman rank correlation test was used to determine statistical correlations. Statistical significance was set at P < 0.05.
3 RESULTS
3.1 METTL3-mediated m6A modification promotes GC malignant progression
It has been conclusively demonstrated that METTL3 expression level was higher in GC tissues than in the adjacent normal gastric mucosa in the TCGA database and clinical patient samples. Elevated METTL3 levels were associated with worse long-term outcomes in GC patients [17]. To examine whether the elevated METTL3 proteins caused the outcome deterioration, we first detected the basic METTL3 protein levels in GC cell lines (Supplementary Figure S1A) and selected the AGS and SGC7901 cell lines with low METTL3 expression to construct METTL3 overexpressing cell models (Supplementary Figure S1B). The proliferation, colony formation, migration, and invasion abilities were significantly improved by METTL3 overexpression (Supplementary Figure S1C-E). In contrast, knockdown of METTL3 (Supplementary Figure S2A) significantly suppressed the proliferation, colony formation, migration, and invasion abilities of AGS, MKN1, and MGC803 cells (Supplementary Figure S2B-D). Considering the m6A catalytic ability and translation enhancement effect of METTL3 protein, we used plasmids containing catalytically inhibited METTL3 (aa395-398, DPPW→APPA), as previously reported [17], to transfected GC cells (Supplementary Figure S3A and B). The m6A dot blot results showed that the catalytic mutant did not affect the total RNA m6A modification level. At the same time, overexpression of wild type (WT) or knockdown of METTL3 significantly influenced RNA m6A levels (Supplementary Figure S3C). Moreover, catalytically inhibited METTL3 abolished METTL3 promoting proliferation, colony formation, migration, and invasion abilities (Supplementary Figure S3D-F). STM2457, a specific inhibitor of METTL3 transmethylation function, also impaired METTL3-mediated proliferation, colony formation, migration, and invasion (Supplementary Figure S4). Collectively, these results suggested that METTL3 promoted GC proliferation, colony formation, migration, and invasion in an m6A-dependent manner.
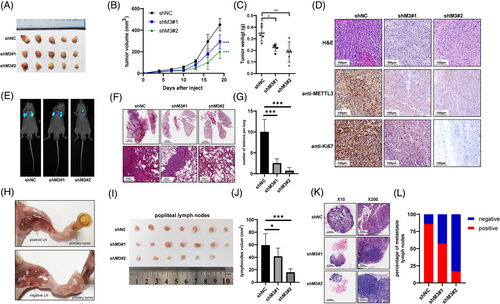
Furthermore, the effect of METTL3 on proliferation and metastasis in vivo was assessed. MGC803 cells treated with METTL3 shRNAs (shM3#1 and shM3#2) or control shRNAs (shNC) were employed. Compared to the negative control, in xenograft models, cells with METTL3 knockdown displayed significantly suppressed tumor growth, both in tumor volume and weight (Figure 1A-C, Supplementary Figure S5). IHC images showed that the proliferation biomarker Ki67 was decreased upon METTL3 knockdown (Figure 1D). Additionally, knockdown of METTL3 significantly inhibited lung metastasis in vivo, as reflected by bioluminescence imaging and H&E staining (Figure 1E-G). As lymph node metastasis is the most common metastasis in GC, we established a footpad-popliteal lymph node metastasis model to investigate the function of METTL3 in lymph node metastasis (Figure 1H). Post harvesting the popliteal lymph nodes after 8 weeks, the lymph nodes from the mice with knockdown of METTL3 cells were smaller than those from negative controls (Figure 1I and J). H&E staining revealed that lymph nodes of METTL3 knockdown groups had a lower percentage of GC metastasis (Figure 1K and L). These results suggested that METTL3 promoted GC proliferation and invasion both in vitro and in vivo.
3.2 METTL3 regulates PBX1 mRNA stability through m6A modification
To explore the molecular mechanism of METTL3 in GC progression, we conducted MeRIP-seq combined with RNA-seq. The MeRIP-seq showed that almost 50% of m6A modification was located in the three prime untranslated regions (3′-UTR) region and 25% in the five prime untranslated regions (5'-UTR; Supplementary Figure S6A and B). Most of these peaks were read both in METTL3 knockdown and negative groups (negative control: 35133; METTL3 knockdown: 33134). And the peaks were mapped in 11755 and 11382 genes respectively (Supplementary Figure S6C). Further analysis revealed that 604 m6A transcripts were lost, and 123 peaks were gained significantly after METTL3 knockdown (Supplementary Figure S6D). Since METTL3 is an m6A writer, we focused on analyzing the genes with lost peaks on METTL3 knockdown. On the other hand, RNA-seq revealed that the expression of 969 genes changed more than 2-fold after the knockdown of METTL3 in AGS cells (Supplementary Figure S6E). We then combined the results of the two sequencing analyses and found that 7 genes overlapped (ZNF264, SMUG1, ZNF668, ZBTB43, ZNF445, PBX1, and ZNF324B; Figure 2A, Supplementary Table S3).
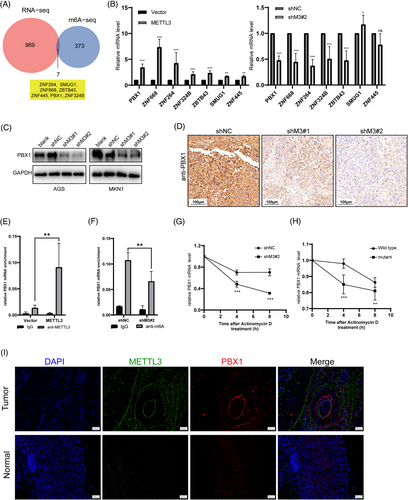
To validate the downstream gene candidates, we used qPCR to measure the mRNA levels of the 7 genes mentioned above in METTL3 (both WT and mutant) overexpressed cells, METTL3 knockdown cells, and corresponding control cells. The results indicated that only ZNF668 and PBX1 transcription levels were significantly downregulated upon METTL3 knockdown and upregulated by elevated METTL3 levels but not influenced by mutant METTL3 levels (Figure 2B, Supplementary Figure S7A). Consistently, the protein level of PBX1 was positively correlated with METTL3 levels in GC cell lines (Figure 2C, Supplementary Figure S7B), whereas mutant METTL3 did not affect PBX1 levels (Supplementary Figure S7C). Moreover, IHC staining of our previous xenograft tumor samples showed that PBX1 expression was inhibited by METTL3 knockdown (Figure 2D). However, protein levels of ZNF668 decreased only in MKN1 cells upon METTL3 overexpression and increased only in SGC7901 cells upon METTL3 knockdown (Supplementary Figure S7D and E). TCGA analysis also confirmed a positive correlation between METTL3 and PBX1 in GC tissues (Supplementary Figure S7F). These data suggested that PBX1, but not ZNF668, may be directly regulated by METTL3.
MeRIP-seq revealed two m6A peaks on the 5'UTR and 3'UTR of PBX1 mRNA that decreased upon METTL3 knockdown (Supplementary Figure S7G). Furthermore, MeRIP analysis showed that PBX1 mRNA was significantly enriched by METTL3 antibody, and the enrichment was increased upon METTL3 overexpression (Figure 2E). The MeRIP-qPCR analysis indicated that the m6A-specific antibody significantly enriched the PBX1 mRNA, and the m6A-specific enrichment was decreased significantly in METTL3-knockdown AGS cells (Figure 2F). We employed actinomycin D to block the RNA transcript of AGS cells as m6A could regulate the stability of RNA targets. The PBX1 mRNA degraded more rapidly upon METTL3 knockdown and was highly stable in METTL3wt-overexpressing AGS cells than those expressing METTL3mut (Figure 2G and H). The IF images of GC tumor and paired adjuvant normal samples confirmed that METTL3 and PBX1 had higher levels in GC tissues than those in normal adjuvant tissues (Figure 2I). We also investigated whether other m6A enzymes affected PBX1 levels. However, there was no variation in PBX1 protein levels in ALKBH5 or METTL14 knockdown (Supplementary Figure S7H and I). These findings indicated that METTL3-mediated m6A modification regulated PBX1 levels by affecting PBX1 mRNA stability.
3.3 METTL3 promotes GC malignant process by elevating PBX1 expression
Both PBX1 and ZNF668 are characteristic oncogenes in some cancer types, but the biological functions of these two genes have not been determined in GC. We explored the RNA profile data of GC patients in the TCGA database. The statistical results showed that patients with higher expression levels of PBX1 or ZNF668 had worse overall and progression-free survival (Figure 3A, Supplementary Figure S8A and B). However, we combined the RNA profile data in TCGA and GTEx databases and found no differences between tumor and normal tissues on PBX1 and ZNF668 mRNA levels (Supplementary Figure S8C). Next, we used two specific siRNAs to inhibit PBX1 and ZNF668 in AGS cell lines. On PBX1 knockdown (Supplementary Figure S8D and E), proliferation viability was significantly suppressed (Figure 3B), and migration and invasion abilities were severely impaired (Figure 3C, Supplementary Figure S8F). However, the knockdown of ZNF668 had the opposite effect on the proliferation, migration, and invasion ability of AGS cells (Supplementary Figure S8G-I). These results suggested that PBX1, but not ZNF668, was a potential oncogene in GC and may be a downstream gene of METTL3 in GC progression.
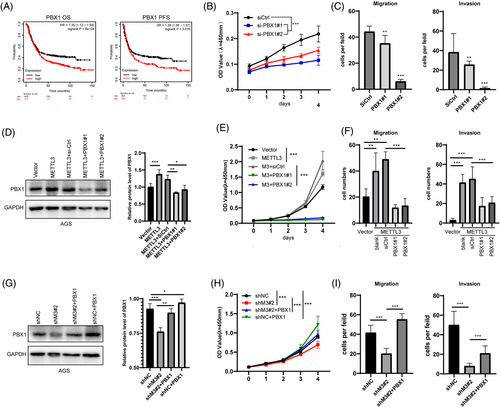
Moreover, we used siRNAs to target PBX1 in AGS and MKN1 cells overexpressing METTL3 and quantified PBX1 protein levels by western blotting (Figure 3D, Supplementary Figure S9A). Knockdown of PBX1 dramatically inhibited the elevation of proliferation, migration, and invasion ability promoted by METTL3 overexpression (Figure 3E and F, Supplementary Figure S9B-D). Correspondingly, overexpression of PBX1 rescued the proliferation, migration, and invasion ability of METTL3-knockdown AGS cells (Figure 3G-I, Supplementary Figure S9E). These results indicated that PBX1 was a target gene of METTL3 that regulated the malignant process of GC.
3.4 PBX1 acts as a TF to induce GCH1 expression in GC
To comprehensively understand the function of PBX1 in GC development, we performed RNA-seq analysis in PBX1-overexpressing and negative control AGS cells, to explore the different genes. The data revealed that a series of pathways related to proliferation and metastasis, such as cell cycle, microtubule-based process, and DNA replication, were remarkably altered upon PBX1 overexpression (Supplementary Figure S10A and B). The molecular mechanism by which PBX1 promoted GC development was then investigated. Given that PBX1 acts as a TF in some cancer types, such as colorectal cancer [29], breast cancer [30], and acute lymphoblastic leukemia [31], we reviewed the CHIP-seq data of cells from colorectal and breast cancer in the CHIPBase database (http://rna.sysu.edu.cn/chipbase3/index.php) to explore potential downstream genes of PBX1. The detected DNA fragments were mapped onto 103 gene promoter regions in colorectal cancer and 3907 genes in breast cancer (Supplementary Table S4). The Venn diagram showed that 35 genes were enriched in both cancer types (Supplementary Figure S10C).
Furthermore, we compared the expression levels of the 35 genes mentioned above in GC and normal tissues in the TCGA database and found that only 23 genes had higher expression levels in GC (Supplementary Figure S10D). Among these genes, GCH1 was reported to be the first rate-limiting enzyme of BH4 biosynthesis. BH4 controls cell proliferation in liver [32] and colon cancer [33]. Thus, we wondered whether the METTL3-PBX1 axis could regulate GCH1 expression.
Accordingly, we then collected 10 tumor tissues and adjacent normal tissues of patients with GC. Western blotting showed that METTL3, PBX1, and GCH1 protein levels were increased in most tumor tissues (Figure 4A). Next, we determined the relationship between the expression levels of METTL3, PBX1, and GCH1 in GC cell lines. The results showed that METTL3 dramatically induced the mRNA and protein levels of GCH1 in an m6A-dependent manner (Figure 4B, Supplementary Figure S10E). In contrast, knockdown of METTL3 in AGS and MKN1 cells markedly suppressed GCH1 levels (Figure 4C and D). After treatment with specific siRNA of PBX1, GCH1 was significantly decreased (Supplementary Figure S10F). Moreover, the catalytic inhibition of METTL3 by STM2457 significantly decreased PBX1 and GCH1 protein levels in a time-dependent manner (Figure 4E). Clinical specimens demonstrated higher protein levels of METTL3 and GCH1 in GC than those in normal adjuvant tissues (Figure 4F). These results suggested that the METTL3-PBX1 axis induced GCH1 expression in GC.
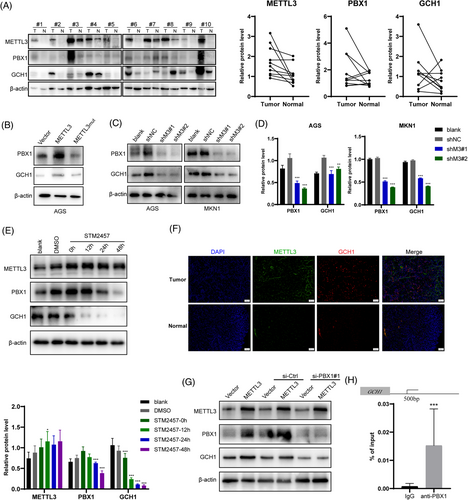
To test whether GCH1 was regulated by PBX1 directly, we used specific siRNAs to block PBX1 expression in AGS cells and found that knockdown of PBX1 eliminated the promoting effect of METTL3 on GCH1 expression (Figure 4G, Supplementary Figure S10G). To explore the transcription mechanism of PBX1, JASPR online tools were used to predict the direct interaction sites between PBX1 and the GCH1 promoter region, which exhibited the likely domain with which the PBX1 protein may combined (Supplementary Figure S10H). Comparing the results from JASPR and CHIPBase, the binding site was suggested to be located approximately 500 bp upstream of the transcription start site (TSS) of GCH1. Next, the ChIP assay indicated that the PBX1 antibody significantly enriched the GCH1 promoter region, while the negative IgG had no such enrichment effect (Figure 4H). These results suggested that PBX1 acted as a TF to induce GCH1 expression, and therefore, GCH1 was the direct downstream gene of the METTL3-PBX1 axis.
3.5 BH4 is the effector molecule of the METTL3-PBX1-GCH1 pathway
As GCH1 was the first rate-limiting enzyme of BH4 biosynthesis [24] (Figure 5A), we measured the BH4 levels in the lysates of cells with overexpression or knockdown of METTL3. BH4 expression was downregulated by METTL3 knockdown and upregulated by METTL3WT overexpression, similar to GCH1 expression; however, it did not change in the cells with METTL3mut transfection (Figure 5B and C). In addition, the knockdown of PBX1 downregulated the intracellular BH4 levels (Figure 5D). The results suggested that METTL3-PBX1 mediated GCH1 protein-regulated BH4 biosynthesis in GC cells.
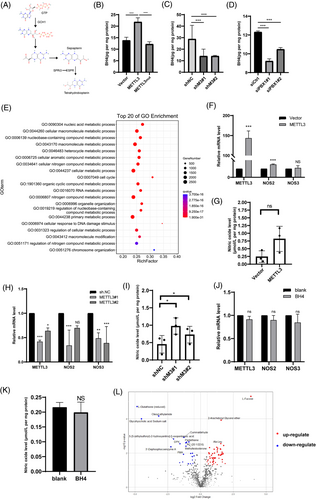
To explore the potential mechanism by which BH4 promotes GC progression, artificial BH4 analogues (sapropterin dihydrochloride) were added to the AGS cell medium. LC-MS showed that the levels of derivatives of BH4, such as biopterin and 7,8-dihydropterin, increased markedly (Supplementary Figure S11A), which conformed to the cell membrane permeability of artificial BH4. RNA-seq revealed that the expression profiles were completely discrepant in BH4-treated cells (Supplementary Figure S11B). GO analysis also showed that the cell cycle and metabolic processes were significantly enriched in BH4-treated cells (Figure 5E, Supplementary Figure S11C). These data suggested that BH4 had a remarkable biological effect on GC cells.
BH4 is a cofactor of nitric oxide synthase (NOS) for NO production. We measured the expression levels of NOS1-3 and NO levels in METTL3 overexpressed or knockdown cells. RT-qPCR revealed that only NOS2 was elevated upon METTL3 overexpression (Figure 5F), whereas NOS1 could not be detected in GC cells (data not shown). However, ELISA of NO production showed that cellular NO levels were unchanged (Figure 5G). Knockdown of METTL3 led to a decrease in NOS2-3, while NO levels slightly increased (Figure 5H and I) and BH4 did not affect NOS or NO levels (Figure 5J and K). These results suggested that BH4 could not regulate GC progression through NO synthases. Previous studies have shown that BH4 regulates cellular metabolic processes depending on its antioxidant characteristics [24]. Therefore, untargeted metabolomic sequencing was performed to verify the metabolism variance in BH4-treated AGS cells. A total of 1097 metabolites were annotated and catalyzed into different categories, such as benzenoids, nucleosides, nucleotides, analogues, lipids, and lipid-like molecules (Supplementary Figure S11D). It was shown that 22 of the mapped metabolites were downregulated and 49 were upregulated upon BH4 treatment (Figure 5L). Consistent with the GO analysis of RNA-seq, the different metabolites included some types of nucleic acids and aromatic amino acids. However, the identification of metabolite functions is still incomplete, and our group will continue to investigate the significance of these metabolites in the future. Collectively, these data suggested that BH4 was the effector molecule of the METTL3-PBX1-GCH1 axis.
3.6 METTL3-PBX1-GCH1 axis increases BH4-induced GC progression
Finally, we investigated whether BH4 promoted GC progression in vitro and in vivo. According to the CCK-8 and transwell analysis, the proliferation, migration, and invasion abilities of AGS cells were dramatically enhanced by BH4 supplementation and restored the proliferation and metastasis defects caused by METTL3 knockdown (Figure 6A and B, Supplementary Figure S12A). In contrast, SPRi3, a small-molecule inhibitor of BH4 synthesis, severely impaired cell proliferation, migration, and invasion abilities (Supplementary Figure S12B and C) and abolished the promoting effect of proliferation, migration, and invasion caused by METTL3 overexpression (Figure 6C and D, Supplementary Figure S12D). A xenograft model was used to verify the suppressive effect of SPRi3 on proliferation. The results showed that SPRi3 abolished the promoting effect of METTL3 on tumor growth (Figure 6E and F, Supplementary Figure S12E). In summary, BH4 biosynthesis, induced via the METTL3-PBX1-GCH1 axis, promoted GC progression in vitro and in vivo.
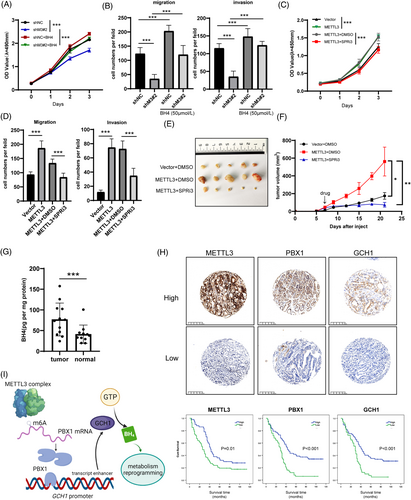
To determine the mechanism in real GC patients, we measured BH4 levels and found the levels to be significantly higher in GC than in normal adjuvant tissues (Figure 6G). We also collected tumor specimens from 117 GC patients at our clinical center with complete follow-up for 5 years. IHC staining was performed on METTL3, PBX1, and GCH1 proteins, and Kaplan-Meier analysis confirmed that these genes indicated a poorer OS of the patients (Figure 6H). Pearson tests revealed that there were positive correlations between METTL3, PBX1, and GCH1 (METTL3 vs. PBX1, r = 0.278, P = 0.0024; METTL3 vs. GCH1, r = 0.175, P = 0.05; PBX1 vs. GCH1, r = 0.38, P <0.001). These data suggested that the METTL3-PBX1-GCH1 axis had worse outcomes in GC patients.
4 DISCUSSION
METTL3 is the most critical methyltransferase protein and forms a catalytic compound core with METTL14 [34]. Overexpression of METTL3 has been reported in various cancer types that promote cancer progression, initiation, and drug resistance [35-37]. In contrast, evidence has shown that METTL3 acts as a cancer suppressor in glioblastoma and bladder cancer [38, 39]. Furthermore, the demethylation proteins ALKBH5 and FTO have oncogene and anti-oncogene functions in some cancer cases [40-45]. These studies indicate that although METTL3 and ALKBH5/FTO oppositely regulate the m6A modification abundance of RNAs, the biological significance of m6A is much more dependent on the effector genes themselves. In GC tissues, Wang et al. [17] showed that METTL3 stabilizes HDGF mRNA to regulate energy metabolism by activating GLUT4 and NEO2. Other genes, such as ZMYM1 and SEC62, have also been identified as target genes regulated by METTL3, promoting GC progression [20, 46]. However, all these findings had no further exploration for clinical practice, because of the absence of specific inhibitors of the downstream pathway. On the other hand, STM2457 has been reported to be a molecular inhibitor of METTL3 [25]. Identifying METTL3-mediated genes is vital for patient selection with STM2457 application in clinical practice. In the present study, we identified the promoting effect of METTL3 on GC progression both in vitro and in vivo. Mechanistically, METTL3-mediated m6A modification stabilized PBX1 mRNA expression. PBX1, having elevated levels, acts as a TF that induces transcription of GCH1, the first rate-limiting enzyme in BH4 biosynthesis, and increases BH4 levels in GC cells and promotes GC progression (Figure 6I). Remarkably, BH4 synthesis had a specific inhibitor, SPRi3, and the anti-tumor effect of the inhibitor had been identified by our group in vivo.
A previous study reported that PBX1 levels increased in the GC mucosa after H. pylori infection [12]. Increased PBX1 level was reported to negatively regulate Cx32 expression, which is associated with H. pylori-related gastric carcinogenesis, indicating that PBX1 may have a potential role in GC oncogenesis [47]. Additionally, the interacting protein of PBX1, HPIP, can induce the cell cycle and Epithelial-mesenchymal transition (EMT) pathway in GC cells and promote GC progression [48]. However, the molecular mechanism of PBX1 overexpression and its oncogenic function remain unclear. The present study found that PBX1 mRNA is stabilized by METTL3-mediated m6A modification. We identified PBX1 as a new oncogene that promotes the proliferation and metastasis of GC cells. In vitro and in vivo studies have indicated that METTL3-mediated PBX1 expression regulates GC proliferation and metastasis. Subsequently, as PBX1 is a well-known TF, we used ChIP-qPCR analysis to confirm the interaction between PBX1 and the GCH1 promoter region. The clinical tissues and cell samples showed a positive correlation between METTL3, PBX1, and GCH1 protein levels.
GCH1 is the first rate-limiting enzyme in BH4 biosynthesis [24]. BH4 acts as a cofactor of enzymes affecting the production of monoamine neurotransmitters and nitric oxide and enzymes affecting pain conduction [21, 22]. Recent studies have shown that BH4 has biological significance in cancer immune reactions and malignant progression [23, 24, 49]. Kynurenine levels are elevated in the GC initiation process, but its downstream metabolite, xanthurenic acid, is a potent inhibitor of a terminal enzyme in the synthetic pathway of BH4 [50]; ELISA analysis of GC samples from our clinical center revealed that BH4 level was increased in tumor tissues. Artificial BH4 administration improved proliferation and metastasis in vitro, while administration of SPRi3 (an inhibitor of BH4 synthesis) had the opposite effect. These results indicate that the m6A-mediated PBX1-GCH1 axis regulates BH4 biosynthesis in GC cells, leading to a malignant process. According to our findings, endocellular NO levels were difficult to detect, and METTL3 did not affect NOS levels.
Furthermore, nitric oxide (NO) levels did not change after BH4 or SPRi3 treatment. Collectively, the promoting effect of BH4 in GC may not depend on the NO synthesis pathway. In contrast, BH4 can work as an antioxidant that influences lipid metabolism or participates in the one-carbon metabolism pathway [24, 51]. Finally, we performed an untargeted metabolomics sequence analysis to preliminarily explore metabolite transformation in BH4 treatment. The data indicated that upon the addition of BH4, the abundance of metabolites, including lipids, amino acids, and nucleotides, was altered in GC cells.
Our study reveals the promoting effect of PBX1 and BH4 in GC development and proposes the m6A-mediated PBX1-GCH1 pathway to regulate BH4 synthesis. Moreover, we suggest that STM2457, an inhibitor of METTL3, and SPRi3, which impairs BH4 biosynthesis, may be valuable in GC therapy. However, there is still a lack of evidence in this study. First, we did not detect the safety and efficiency of STM2457 or SPRi3 in vivo. Second, the mechanism and biological significance of metabolism remodeling induced by BH4 are yet to be explored in our subsequent studies.
5 CONCLUSIONS
Our data revealed that METTL3 regulates the expression of potential oncogene PBX1 in an m6A-dependent manner. Moreover, PBX1 plays a role in elevating GCH1 expression. The METTL3-PBX1-GCH1 axis controls the synthesis of BH4 and promotes the proliferation, migration, and invasion of GC cells. Therefore, targeting the METTL3-PBX1-GCH1 axis may be a potential strategy for facilitating GC-targeted therapy.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Institutional Ethical Boards of the First Affiliated Hospital of Sun Yat-sen University. Written informed consent was obtained from all patients. The animal study was carried out in compliance with the guidance suggestion of Animal Care Committee of the First Affiliated Hospital of Sun Yat-sen University (permit number: [2021]737).
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This study was supported by the research grants from the National Natural Science Foundation of China (81702878, 82003112, and 81372341), Guangdong Basic and Applied Basic Research Foundation (2020A1515010214, 2018A030313634, and 2021A1515010473), Project 5010 of The First Affiliated Hospital of Sun Yat-sen University (2018004), Young Teacher Training Project of The First Affiliated Hospital of Sun Yat-sen University (19ykpy58), and Guangzhou Basic and Applied Basic Research Foundation (202102080538) and China Postdoctoral Science Foundation (2020M683087).
AUTHORS' CONTRIBUTIONS
Ertao Zhai designed the study. Yinan Liu and Ertao Zhai contributed equally to the experiment implement, analysis and interpretation of data, and critical revision of the manuscript for important intellectual content. Junting Chen, Yan Qian, Risheng Zhao, Yan Ma, Jianqiu Liu, and Zhixin Huang participated in the experiment, sample collection, and statistical analysis. Yinan Liu wrote the whole paper. Jianhui Chen and Shirong Cai supervised the whole study. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
The authors thank YiJin (Guangzhou, Guangdong, China) for plasmids construction and LC-Bio Technologies Co., Ltd (Hangzhou, Zhejiang, China) for the support of Me-RIP and RNA-seq. We thank to Novogene Co., Ltd (Beijing, China) for the technology of untargeted sequence of metabolism.




