Genetic alternations and immune characteristics in patients with small cell lung cancer
Abbreviations
-
- AES
-
- Amino-Terminal Enhancer Of Split
-
- AXIN1
-
- Axis Inhibition Protein 1
-
- BCL9
-
- B Cell CLL/Lymphoma 9
-
- BMP5
-
- Bone Morphogenetic Protein 5
-
- BWA
-
- Burrows-Wheeler Aligner
-
- C8orf82
-
- Chromosome 8 Open Reading Frame 82
-
- CALR
-
- Calreticulin
-
- CBWD3
-
- COBW Domain Containing 3
-
- CBX4
-
- Chromobox 4
-
- CLPTM1L
-
- Cleft Lip And Palate Transmembrane Protein 1-Like Protein
-
- CMBL
-
- Carboxymethylenebutenolidase Homolog
-
- CNVs
-
- copy number variations
-
- COL11A1
-
- Collagen Type XI Alpha 1 Chain
-
- CPS
-
- combined positive score
-
- CREBBP
-
- CREB Binding Protein
-
- CSMD1
-
- CUB And Sushi Multiple Domains 1; CSMD3, CUB And Sushi Multiple Domains 3
-
- CTNNA2
-
- Catenin Alpha 2
-
- CYP2C8
-
- Cytochrome P450 Family 2 Subfamily C Member 8
-
- CYSLTR2
-
- Cysteinyl Leukotriene Receptor 2
-
- DAB
-
- diaminobenzidine
-
- DAXX
-
- Death Domain Associated Protein
-
- DNAH5
-
- Dynein Axonemal Heavy Chain 5
-
- ELPS
-
- Enhance Labelled Polymer System
-
- EPHA7
-
- Ephrin Type-A Receptor 7
-
- ERBB2
-
- Erb-B2 Receptor Tyrosine Kinase 2
-
- ERCC4
-
- Excision Repair Cross-Complementation Group 4
-
- ERICH3
-
- Glutamate Rich 3
-
- ERRFI1
-
- ERBB Receptor Feedback Inhibitor 1
-
- FAM131A
-
- Family With Sequence Similarity 131 Member A
-
- FAT2
-
- FAT Atypical Cadherin 2
-
- FDR
-
- false discovery rate
-
- FFPE
-
- formalin-fixed and parrffin-embedded
-
- GADD45G
-
- Growth Arrest And DNA Damage Inducible Gamma
-
- GATK
-
- Genome Analysis Toolkit
-
- GISTIC
-
- Genome Identification of Significant Targets in Cancer
-
- GOLGA6L6
-
- Golgin A6 Family Like 6
-
- GRB2
-
- Growth Factor Receptor Bound Protein 2
-
- HES 1
-
- Hes Family BHLH Transcription Factor 1
-
- HES6
-
- Hes Family BHLH Transcription Factor 6
-
- HLTF
-
- Helicase Like Transcription Factor
-
- HMGN2
-
- High Mobility Group Nucleosomal Binding Domain 2
-
- InDels
-
- insertions and deletions
-
- ISX
-
- Intestine Specific Homeobox
-
- KCNC3
-
- Potassium Voltage-Gated Channel Subfamily C Member 3
-
- KLF6
-
- Kruppel Like Factor 6
-
- KMT2D
-
- Lysine Methyltransferase 2D
-
- LRIG3
-
- Leucine Rich Repeats And Immunoglobulin Like Domains 3; LRP1B, LDL Receptor Related Protein 1B
-
- LRP2
-
- Low-Density Lipoprotein Receptor-Related Protein 2
-
- MACC1
-
- Metastasis Associated In Colon Cancer 1
-
- MAZ
-
- MYC Associated Zinc Finger Protein
-
- MBD3
-
- Methyl-CpG Binding Domain Protein 3
-
- MIDN
-
- Midnolin
-
- MSH6
-
- MutS Homolog 6
-
- MUC16
-
- Mucin 16, Cell Surface Associated; MUC17, Mucin 17, Cell Surface Associated
-
- MXI1
-
- MAX Interactor 1, Dimerization Protein
-
- NAV3
-
- Neuron Navigator 3
-
- NDUFS6
-
- NADH:Ubiquinone Oxidoreductase Subunit S6
-
- NKX2-1
-
- NK2 Homeobox 1
-
- NMF
-
- non-negative matrix factorization
-
- NOTCH1
-
- Notch Homolog 1; NOTCH3, Notch Receptor 3
-
- NPIPA1
-
- Nuclear Pore Complex Interacting Protein Family Member A1
-
- NPRL2
-
- NPR2 Like, GATOR1 Complex Subunit
-
- NTHL1
-
- Nth Like DNA Glycosylase 1
-
- OS
-
- overall survival
-
- OS
-
- overall survival
-
- PARP3
-
- Poly(ADP-Ribose) Polymerase Family Member 3
-
- PBS
-
- phosphate buffered saline
-
- PD-L1
-
- programmed cell death-ligand 1
-
- POLE
-
- DNA Polymerase Epsilon, Catalytic Subunit
-
- POLH
-
- DNA Polymerase Eta
-
- POTEJ
-
- POTE Ankyrin Domain Family Member J
-
- PPP2R1A
-
- Protein Phosphatase 2 Scaffold Subunit Aalpha
-
- PTEN
-
- Phosphatase And Tensin Homolog
-
- PTMS
-
- Parathymosin
-
- PTPN6
-
- Protein Tyrosine Phosphatase Non-Receptor Type 6
-
- RAF1
-
- Raf-1 Proto-Oncogene, Serine/Threonine Kinase
-
- RB1
-
- retinoblastoma 1
-
- REV1
-
- REV1 DNA Directed Polymerase
-
- ROBO2
-
- Roundabout Homolog 2
-
- RYR2
-
- Ryanodine Receptor 2
-
- SCLC
-
- small cell lung cancer
-
- SIRPA
-
- Signal Regulatory Protein Alpha
-
- SLC9A3
-
- Solute Carrier Family 9 Member A3
-
- SNVs
-
- single nucleotide variations
-
- SPRED1
-
- Sprouty Related EVH1 Domain Containing 1
-
- SYNE1
-
- Spectrin Repeat Containing Nuclear Envelope Protein 1
-
- TBC1D3
-
- TBC1 Domain Family Member 3
-
- TERT
-
- Telomerase Reverse Transcriptase
-
- TIMELESS
-
- Timeless Circadian Regulator
-
- TMB
-
- tumor mutational burden
-
- TP53
-
- tumor protein p53
-
- TPPP
-
- Tubulin Polymerization Promoting Protein
-
- TPS
-
- tumor proportion score
-
- TPSAB1
-
- Tryptase Alpha/Beta 1
-
- TRAF7
-
- TNF Receptor Associated Factor 7
-
- TRIM49C
-
- Tripartite Motif Containing 49C
-
- TSC2
-
- Tuberous Sclerosis 2
-
- TSPY1
-
- Testis Specific Protein Y-Linked 1
-
- UBL4A
-
- Ubiquitin Like 4A
-
- USH2A
-
- Usher Syndrome 2A
-
- WES
-
- whole exome sequencing
-
- WES
-
- whole-exome sequencing
-
- XIRP2
-
- Xin Actin Binding Repeat Containing 2
-
- ZFHX4
-
- Zinc Finger Homeobox 4
-
- ZNF479
-
- Zinc Finger Protein 479
Dear Editor,
As an aggressive and recalcitrant subtype of lung cancer, small cell lung cancer (SCLC) is linked with a dismal prognosis where chemotherapy remains the backbone of treatment. In this disappointing context, immunotherapy has brought hope for patients with SCLC [1]. However, data on the genomic and immunological landscape of SCLC are urgently needed to achieve more precise and effective treatment. Here, we conducted a comprehensive analysis of genetic alteration and immune characteristics in a cohort of Chinese patients with SCLC.
Whole-exome sequencing (WES) was performed to identify gene mutations and copy number variations (CNVs) among 178 SCLC patients. The data from WES were deposited to Genome Sequence Archive (http://bigd.big.ac.cn/gsa or http://gsa.big.ac.cn) in Data Center of Beijing Institute of Genomics under the accession number subHRA001430. Immunohistochemical staining was conducted to evaluate programmed cell death-ligand 1 (PD-L1) expression and CD8+ T cell infiltration. Details of all procedures can be found in the Supplementary Materials and Methods. Among these 178 SCLC patients (median age, 62 years; range of age, 29-79 years), 86 (48.3%) patients were at limited stage and 92 (51.7%) were at extensive stage. The clinicopathological characteristics of the patients are presented in Supplementary Table S1.
The gene mutation and CNV landscapes of these patients were characterized by gender, smoking status, family history, drinking status, stage, and age. We showed that the top ten frequently mutated genes were tumor protein p53 (TP53) (93.3%), retinoblastoma 1 (RB1) (44.4%), Notch receptor 1 (NOTCH1) (20.2%), CREB binding protein (CREBBP) (18.0%), FAT atypical Cadherin 2 (FAT2) (12.9%), helicase like transcription factor (HLTF) (7.9%), protein phosphatase 2 scaffold subunit aalpha (PPP2R1A) (6.7%), phosphatase and tensin homolog (PTEN) (6.2%), Erb-B2 receptor tyrosine kinase 2 (ERBB2) (5.6%), NPR2 like, GATOR1 complex subunit (NPRL2) (5.1%) (Figure 1A). Genes frequently altered in SCLC reported in other studies [2, 3], such as TP53, RB1, PTEN, ERBB2 and CREBBP, were seen in our cohort. In George et al.’s study [3], RB1 was altered in all but two cases that exhibited signs of chromothripsis, which indicates a heterogeneous genomic landscape among different ethnicities. We further compared our cohort with another Chinese cohort [4]. The frequency of RB1 mutation was 44.4% in our SCLC cohort and was 62.0% in Jiang et al.’s study [4]. Reasons for this discrepancy could be the differences in the proportion of patients at extensive stage (52.0% vs. 4.0%), the proportion of inclusion of controls in WES (100.0% vs. 25.0%), and the percentage of patients undergoing treatments (0.0% vs 9.0%).
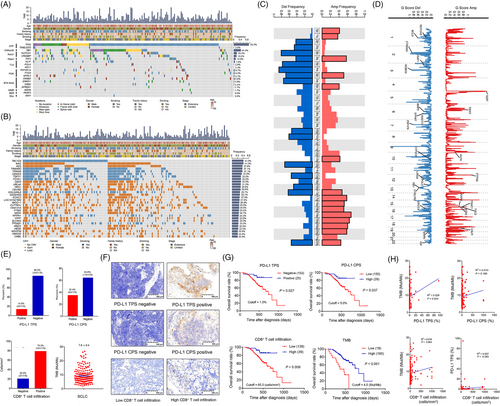
Furthermore, the associations between the top 20 frequently mutated genes and immunological profiles have been analyzed. Significant differences in CD8+ T cell infiltration were found between patients with Usher syndrome 2A (USH2A), CUB and Sushi multiple domains 1 (CSMD1), Notch Receptor 3 (NOTCH3) mutations and those with these wild-type genes (all P < 0.05) (Supplementary Figure S1). Significant differences were also detected in PD-L1 tumor proportion score (TPS) between the SCLC patients harboring Mucin 16, cell surface associated (MUC16), USH2A, spectrin repeat containing nuclear envelope protein 1 (SYNE1), low-density lipoprotein receptor-related protein 2 (LRP2) mutations and their wildtype (all P < 0.05) (Supplementary Figure S2). Significant differences in PD-L1 combined positive score (CPS) were found in MUC16-mutant and USH2A-mutant SCLC patients compared with their wild-type (both P < 0.01) (Supplementary Figure S3). The tumor mutation burden (TMB) was significantly higher in patients harboring MUC16, ryanodine receptor 2 (RYR2), zinc finger homeobox 4 (ZFHX4), USH2A, CUB And Sushi Multiple Domains 3 (CSMD3), LDL Receptor Related Protein 1B (LRP1B), Mucin 17, Cell Surface Associated (MUC17), dynein axonemal heavy chain 5 (DNAH5), SYNE1, xin actin binding repeat containing 2 (XIRP2), glutamate rich 3 (ERICH3), lysine methyltransferase 2D (KMT2D), LRP2, collagen type XI alpha 1 chain (COL11A1) and NOTCH3 mutations than in those harboring wildtype genes (all P < 0.05) (Supplementary Figure S4).
The top ten genes with most frequent CNVs were TBC1 Domain Family Member 3 (TBC1D3) (41.0%), MYC associated zinc finger protein (MAZ) (41.0%), amino-terminal enhancer of split (AES) (37.6%), tripartite motif containing 49C (TRIM49C) (37.1%), TRIM49 (37.1%), COBW domain containing 3 (CBWD3) (34.8%), testis specific protein Y-linked 1 (TSPY1) (32.0%), POTE ankyrin domain family member J (POTEJ) (32.0%), chromobox 4 (CBX4) (32.0%), ubiquitin like 4A (UBL4A) (31.5%) (Figure 1B). Interestingly, the genetic CNVs were demonstrated in either full gain or full loss. We only listed the top 30 genes with the most frequent CNVs (Figure 1B). In addition, Myc family member amplification, reported in 14.0%-18.0% of SCLC cases [3, 4], occurred in 12.9% of our cohort.
At the chromosomal arm level, significant amplification of chromosomal 1p, 1q, 3q, 5p, 6q, 12q, 17p, 17q, 18p, 18q, 19p, 19q, 20p, 20q, 22q and deletion of chromosomal 2p, 2q, 3p, 3q, 4p, 4q, 5p, 5q, 10p, 10q, 13q, 15q, 16q, 17p, 22q were detected in our SCLC cohort (Figure 1C). At the focal level, significant amplification and deletion of several genes were also observed (Figure 1D).
In the present study, 25 (14.0%) patients were PD-L1 TPS-positive and 63 (35.4%) were PD-L1 CPS-positive (Figure 1E). However, only 3.0% were PD-L1 TPS-positive in Chen's study [5], which could be possibly ascribed to the differences in stage, cutoff for PD-L1, and PD-L1 antibodies. We also found that 37 (20.8%) patients had no CD8+ T cell infiltration. The median TMB was 7.6 Mut/Mb (Figure 1E). Representative immunohistochemical staining of PD-L1 and CD8+ T cell infiltration are demonstrated in Figure 1F.
We further showed that positive PD-L1 TPS and high expression were associated with prolonged survival (Figure 1G) whereas positive PD-L1 CPS was not (Supplementary Figure S5). The prognostic role of CD8+ T cell infiltration in SCLC is controversial [6, 7]. In our study, CD8+ T cell infiltration was a positive prognostic factor affecting OS (P = 0.008) (Figure 1G). Interestingly, high TMB was found to be associated with improved OS (P < 0.001) (Figure 1G). Additionally, a positive correlation between PD-L1 TPS and TMB was found (R2 = 0.029, P = 0.024), while no significant correlations between PD-L1 CPS and TMB, between CD8+ T cell infiltration and TMB, and between CD8+ T cell infiltration and PD-L1 TPS were observed (Figure 1H).
Nevertheless, several limitations existed in the present study. The biopsy specimens might not be representative of genetic mutation quantification compared to whole sections from larger surgical specimens. WES may identify unknown genetic mutations in a wider range and with more accuracy than whole-genome sequencing. However, it is expensive, complicated and time-consuming, limiting its clinical utility. Other immunological factors such as CD47 and CD45, vital for SCLC tumor microenvironment, were not used in the present study.
In conclusions, we depicted the genomic mutation and CNV profiles of Chinese SCLC patients. Some similarities in genomic features exist between our cohort and other reported cohorts. However, the Chinese cohort has its own unique features, as exemplified by low RB1 mutation rate and distinct CNV landscape. Moreover, we revealed that SCLC patients with high PD-L1 expression, CD8+ T cell infiltration, and TMB may have prolonged survival. These unique features could pave the way for discovering potential therapeutic targets for Chinese SCLC patients.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Ethics Committee of Shandong Cancer Hospital and Institute. All included patients in this study offered written informed consent.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Methods and materials are available in the supplementary file. The data from whole-exome sequencing were deposited to Genome Sequence Archive in Data Center of Beijing Institute of Genomics under the accession number subHRA001430.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This study was supported jointly by Special Funds for Taishan Scholars Project (Grant No. tsqn201812149) and Academic Promotion Program of Shandong First Medical University (2019RC004).
ACKNOWLEDGEMENTS
We thank Dianbin Mu from Shandong Cancer Hospital for his technical support in pathology, and we thank Zhenzhen Li, Shuangxiu Wu and Yanxiang Zhang from Berry Oncology Corporation for their technical support in bioinformatics.
AUTHORS’ CONTRIBUTIONS
CYZ collected the data and wrote the article. CLZ made figures, pathological confirmed and immunohistochemical analysis. HYW carried out the overall design of the research and supervision of the article.




