pH-Dependent Packing Mode Variations and Chirality Inversion in Short Peptide Self-Assembly
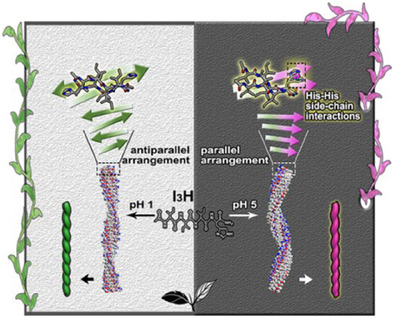
Graphical Abstract
This work reports subtle pH-dependent chirality inversions through designed amphiphilic peptides. The underlying mechanism is revealed to be closely related to the protonation state of the hydrophilic His residue. The protonated His promotes antiparallel β-sheet packing and left-handed fibril assembly, whereas the deprotonated His facilitates parallel packing and right-handed fibril assembly, driven by interstrand His–His interactions.
Abstract
Precise control of structures and morphologies in peptide self-assembly has been challenging. We report the self-assembly of amphiphilic peptides I3H, designed with a modular structure featuring three consecutive isoleucine residues as a hydrophobic tail and a C-terminal histidine-based hydrophilic headgroup. Microscopic, neutron scattering, and spectroscopic techniques demonstrate that the designed peptides self-assemble into β-sheet nanofibrils, with their helix handedness exhibiting subtle pH-dependent inversion. pH titration, NMR, and molecular dynamics simulations reveal the underlying mechanism correlates with the protonation state of histidine and the molecular packing modes in β-sheet assemblies. The protonated histidine promotes antiparallel β-sheet packing at lower pH while its deprotonated state favors parallel packing when pH is increased. Strong π-π stacking interactions between deprotonated histidine side chains in parallel β-sheet arrangements drive chiral flipping of β-strands, ultimately inducing supramolecular helix inversion. Furthermore, such a pH-dependent helix inversion can be engineered by inserting the achiral and flexible glycine at the hydrophobic/hydrophilic interface, with I3GH assembly maintaining this effect while I3GGH assembly abolishing it. This work not only advances our mechanistic understanding of peptide chirality inversion at the level of individual β-sheets but also provides a blueprint for designing hierarchical chirality through precise modulation of molecular packing modes and side-chain interactions.
Conflict of Interests
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information of this article.