The ATM Kinase Inhibitor AZD0156 Is a Potent Inhibitor of Plasmodium Phosphatidylinositol 4-Kinase (PI4Kβ) and Is an Attractive Candidate for Medicinal Chemistry Optimization Against Malaria
Graphical Abstract
AZD0156, an ATM kinase inhibitor in clinical development, shows promising multistage antiplasmodial activity by targeting Plasmodium falciparum phosphatidylinositol 4-kinase (PfPI4Kβ). With an improved specificity profile relative to other PfPI4Kβ inhibitors and moderate efficacy in a P. berghei disease model, AZD0156 is an attractive candidate for medicinal chemistry optimization against malaria.
Abstract
New compounds targeting human malaria parasites are critical for effective malaria control and elimination. Here, we pursued the imidazoquinolinone AZD0156 (MMV1580483), a human ataxia-telangiectasia mutated (ATM) kinase inhibitor that completed Phase I clinical trials as an anticancer agent. We validated its in vitro activity against the two main forms of the Plasmodium falciparum parasite in the human host, viz. the asexual blood (symptomatic) stage and sexual gametocyte (transmission) stage. Resistance selection, cross-resistance, biochemical, and conditional knockdown studies revealed that AZD0156 inhibits P. falciparum phosphatidylinositol 4-kinase type III beta (PfPI4Kβ), a clinically-validated target for the treatment of malaria. Metabolic perturbations, fixed-ratio isobolograms, killing kinetics and morphological evaluation correlated AZD0156 inhibition with other known PI4Kβ inhibitors. The compound showed favorable in vivo pharmacokinetic properties and 81% antimalarial efficacy (4 × 50 mg kg−1) in a P. berghei mouse malaria infection model. Importantly, a cleaner biochemical profile was measured against human kinases (MAP4K4, MINK1) implicated in embryofoetal developmental toxicity associated with the PfPI4Kβ inhibitor MMV390048. This improved kinase selectivity profile and structural differentiation from other PI4Kβ inhibitors, together with its multistage antiplasmodial activity and favorable pharmacokinetic properties, makes AZD0156 an attractive candidate for target-based drug repositioning against malaria via a medicinal chemistry optimization approach.
Introduction
The efficacy of the current arsenal of clinical antimalarials, the cornerstone of parasite control, is constantly threatened by the emergence and spread of drug-resistant strains of the most virulent human malaria parasite, Plasmodium falciparum.[1-3] This calls for the urgent development of new chemotherapies, ideally with novel modes of action and activity against multiple stages of P. falciparum, to circumvent untreatable infections and contribute to malaria elimination.[4, 5] Novel antimalarials that simultaneously target the pathogenic asexual blood stage (ABS) parasites and transmissible gametocytes will have both therapeutic and transmission-blocking relevance, particularly if a vulnerable and essential biological pathway is targeted.[5, 6]
In this respect, due to their essentiality and expression across multiple life cycle stages, Plasmodium kinases are attractive as targets for the development of potential multistage-active antimalarials.[4, 7-9] One such example is the clinically-validated target Plasmodium phosphatidylinositol 4-kinase type III beta (PI4Kβ)[4, 7] which catalyzes the conversion of phosphatidylinositol to phosphatidylinositol 4-phosphate to regulate intracellular signalling and trafficking.[10, 11] MMV390048, an aminopyridine that reached Phase II clinical trials for the treatment of malaria, was validated as a potent Plasmodium PI4Kβ inhibitor.[2] However, embryofoetal studies prevented the further development of MMV390048 as off-target inhibition of the mammalian PI4Kβ orthologue, in combination with the mammalian mitogen-activated protein kinase 4 (MAP4K4) and misshapen-like kinase 1 (MINK1), were speculated to result in developmental toxicity signals in rats. Interestingly, these toxicities were not observed in rabbits.[12] Hence, despite the druggable nature of PfPI4Kβ, there is a pressing need to identify new chemotypes with a cleaner selectivity and toxicity profile.
To leverage existing data and mitigate time and cost risks, the repositioning of anticancer compounds for malaria has recently attracted considerable interest.[13-16] In this context, “repositioning” refers to using an existing compound as a starting point for medicinal chemistry optimization towards treatment for a new indication, and is contrasted with “repurposing” in which a new compound is used directly to treat a different indication than the one for which it was originally developed, without chemical modification of that compound. Parallel screens of the open-source Medicines for Malaria Venture (MMV) Pandemic Response Box (PRB) identified AZD0156 (MMV1580483) as having in vitro antiplasmodial activity across multiple stages of P. falciparum development, viz. the liver stages, ABS parasites and transmissible stages (gametocytes).[17] This imidazoquinolinone was developed as a human ataxia-telangiectasia mutated (ATM) kinase inhibitor and completed Phase 1 clinical trials for advanced solid tumours, alone and in combination with other anticancer treatments (https://clinicaltrials.gov/study/NCT02588105).
Here, we explored the potential of repositioning AZD0156 as a multistage antimalarial candidate through extensive mechanistic evaluation against P. falciparum parasites. Phenotypic approaches and biochemical profiling revealed that AZD0156 inhibits PfPI4Kβ. Although this kinase has been shown to be an essential and promising multistage target,[11] off-target activities due to the conserved kinase ATP binding site have created challenges for developing selective PfPI4Kβ inhibitors. Here we show that AZD0156 has minimal off-target interactions toward other key P. falciparum and human kinases, unlike those previously reported for other PfPI4Kβ inhibitors. This unique selectivity profile, combined with good in vitro and in vivo efficacy, positions this structurally-differentiated chemotype as an attractive starting point for further medicinal chemistry optimization as a potential novel treatment and transmission-blocking agent for malaria.
Results and Discussion
Hit Confirmation of the Multistage Activity of AZD0156
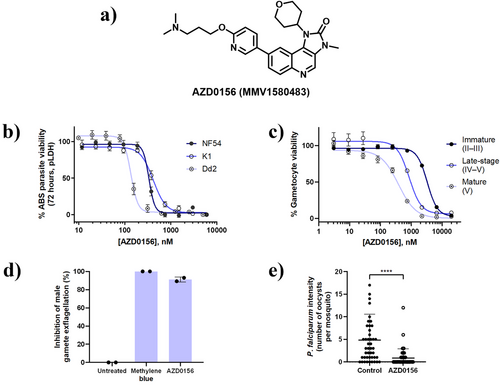
AZD0156 (Figure 1a) was previously identified as a singleton hit after screening the MMV pandemic response box diversity set.[17] The compound was resynthesized and the reported multistage activities were confirmed (Figure 1). AZD0156 displayed submicromolar in vitro antiplasmodial activity against drug-sensitive (PfNF54 IC50 335 ± 23 nM) and drug-resistant (PfK1 IC50 415 ± 23 nM; PfDd2 IC50 136 ± 7 nM) ABS parasites in a 72 h metabolic parasite lactate dehydrogenase (pLDH) assay (Figure 1b). Late-stage gametocyte (>90% stages IV–V) activity on a luciferase-reporter assay (Figure 1c; IC50 943 ± 41 nM) were in alignment with previous data obtained using the PrestoBlue cell viability platform (236 ± 0 nM).[17] The activity against these forms of the parasite was further interrogated on both immature (>90% stages II–III) and mature (>95% stage V) gametocytes. Interestingly, AZD0156 showed a decrease in activity against PfNF54 immature gametocytes (IC50 3 312 ± 76 nM) but, notably, AZD0156 showed equipotent mature gametocyte activity (IC50 384 ± 37 nM) compared to ABS parasites (< two-fold change). The decrease in activity against immature gametocytes is a phenotype rarely observed for small molecules against these gametocyte stages.[18] However, the activity against mature gametocytes prompted the assessment of transmission-blocking potential of AZD0156 against male gamete exflagellation (exflagellation inhibition assay) and oocyst formation (standard membrane feeding assay, SMFA). In line with previously-reported transmission-blocking activity,[17] AZD0156 showed 90% inhibition of male gamete exflagellation at 2 µM (Figure 1d). Additionally, AZD0156 significantly reduced oocyst prevalence with a 56 ± 6% transmission-blocking and 82 ± 5% transmission-reducing activities (Figure 1e), respectively.
Plasmodium PI4Kβ Is the Primary Target of AZD0156
Selecting for resistant mutants against ABS P. falciparum parasites in vitro, followed by whole-genome sequencing, is an unbiased approach to identify resistance mechanisms and potential drug targets. Therefore, resistance selections with AZD0156 were performed by exposing 2 × 109 PfDd2 parasites in two separate culture flasks to AZD0156 at 3 × IC50. After 22 days, a resistant population recrudesced in one flask and the bulk culture was cloned via limiting dilution. The profiled clones, D6 and F3, were ∼three-fold less sensitive to AZD0156 (IC50 389 ± 13 nM and 337 ± 5 nM, respectively) compared with the PfDd2 parental line (IC50 121 ± 13 nM) (Figure 2a). Whole-genome sequencing of clones D6 and F3 revealed a single nucleotide polymorphism in pfpi4kβ (PF3D7_0509800) translating to a S1320L mutation in PfPI4Kβ (Figure S1A). Interestingly, the PfPI4Kβ S1320L mutation was previously observed following a resistance selection with the HsPI4KIIIβ and PfPI4Kβ inhibitor, BQR695.[11] Clone D6 additionally harboured a mutation in a putative trafficking protein (Figure S1A); however, no copy number variations (CNVs) were observed in either clone.
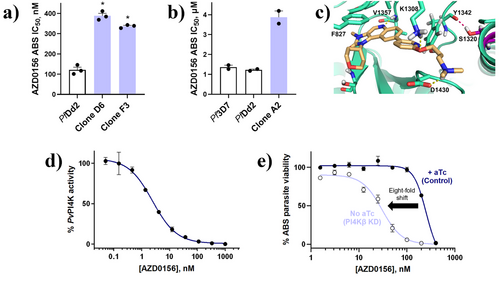
The clinical candidate MMV390048 also targets PfPI4Kβ and resistance against MMV390048 was due to a A1319V mutation in PfPI4Kβ that caused a four-fold IC50 shift for MMV390048 relative to the PfDd2 parental line.[19] Notably, the position of the A1319V mutation in PfPI4Kβ is adjacent to the S1320L mutation selected by AZD0156. This PfPI4Kβ A1319V mutant parasite line (clone A2) was subsequently employed to cross-screen for the potential of AZD0156 resistance, whereupon a differential susceptibility relative to PfDd2 was also observed with an average fold IC50 shift for AZD0156 of 3.2 (Figure 2b), tracking with the phenotypic shifts of the PfPI4Kβ-S1320L mutants.
Based on a PfPI4Kβ homology model,[20] S1320L is located in the ATP-binding site between the hinge binding region and affinity pocket (Figure 2c). Interestingly, the S1320L mutation results in loss of a hydrogen bond that is typically formed in the wild type protein between S1320 and Y1342, and this could distort the binding site in the mutated protein relative to the wild type, giving rise to the resistant phenotype. Molecular docking predictions in the wild type isoform show that AZD0156 forms key interactions with the phosphate-binding loop (P-loop, F827), catalytic site (K1308), hinge-binding region (V1357), ribose pocket (S1362), and affinity pocket (D1430) (Figures 2c and S1B).[20]
To confirm PfPI4Kβ as the target of AZD0156 in vitro, we used a P. vivax PI4Kβ recombinant enzyme assay, based on a 97% sequence homology between the PfPI4K and PvPI4K ATP-binding site and catalytic region and known difficulties for expressing PfPI4Kβ.[15] Against PvPI4Kβ, we recorded an IC50 value of 5 ± 2 nM for AZD0156 (Figure 2d), directly comparable to previously-validated PfPI4Kβ inhibitors MMV390048, UCT943 and sapanisertib, all of which recorded IC50 values <5 nM in this assay.[4, 19, 21]
We also probed the effect of PI4Kβ knockdown on parasite sensitivity to AZD0156 using an engineered PfPI4Kβ conditional knockdown (cKD) parasite line.[15] In this assay, PfPI4Kβ translation is reduced by the removal of anhydrotetracycline (aTc) through the TetR (Tet repressor protein)/DOZI (development of zygote inhibited)-RNA aptamer module. Thus, a low aTc concentration constitutes a conditional PfPI4Kβ knockdown.[22, 23] The cKD of PfPI4Kβ led to eight-fold increased parasite sensitivity to AZD0156 (Figure 2e, IC50 30 ± 2 nM and 252 ± 40 nM, respectively), comparable to a > 10-fold shift previously reported for the PfPI4Kβ inhibitor, sapanisertib.[15] This is consistent with decreased target protein requiring less compound for parasite growth to be inhibited. Thus, in line with observations for other validated PfPI4Kβ inhibitors, these data substantiated PfPI4Kβ as the primary target of AZD0156.
AZD0156 has a Cleaner Selectivity Profile for PfPI4Kβ Compared to Other PI4Kβ Inhibitors
Off-target activity is a significant challenge when developing kinase inhibitors due to the conserved ATP-binding site of this superfamily of enzymes. ATM kinases propagate an extensive signalling cascade in response to DNA damage; hence, ATM kinase inhibitors are frequently used with radiation therapy or other DNA-damaging agents.[24-26] Plasmodium parasites have a divergent cell cycle progression profile evident from a lack of canonical cell cycle checkpoint proteins, such as Rb and p53 proteins, and ATM and ATR kinases.[27, 28] PfPI3K has also been identified as a potential kinase inhibitor target of AZD0156[15] and inhibition data for AZD0156 against the human homologue of PfPI4K, HsPI3K, have already been published (0.32 µM, 1.8 µM, 1.1 µM, and 0.27 µM for the α, β, γ, and δ isoforms of HsPI3K, respectively[26]); however, this did not preclude the compound from entering and completing Phase I clinical development.
In light of a report speculating that PfPI4Kβ inhibitors, specifically MMV390048, may interact deleteriously with mammalian MAP4K4 and MINK1, in combination with HsPI4Kβ, leading to embryofoetal toxicity (teratogenicity),[12] we evaluated biochemical activity against these human kinases. For HsMAP4K4 and HsMINK1, AZD0156 showed <50% enzyme inhibition at 10 µM, compared to nanomolar inhibition by the reference compound, staurosporine, a promiscuous ATP-competitive kinase inhibitor (Figure 3a,b). This was a promising improvement on the inhibition data previously measured for MMV390048, viz. 0.8 µM and 0.7 µM against HsMAP4K4 and HsMINK1, respectively.[12] Although AZD0156 showed partial inhibition of the PfPI4Kβ orthologue, HsPI4Kβ (IC50 1.22 µM, Figure 3c), which is comparable to that previously reported for MMV390048 (1 µM),[12] these are encouraging data which indicate that the imidazoquinolinone scaffold has a biochemical profile that is distinct from other PfPI4K inhibitors. This improved selectivity profile provides an attractive starting point for medicinal chemistry optimization of this chemotype.
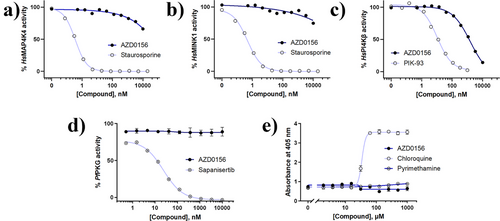
Recent work has demonstrated that PfPI4Kβ inhibitors may also target other enzymes or pathways in P. falciparum in a dual inhibitor fashion; for example, the anticancer kinase inhibitor sapanisertib targets both PI4Kβ and P. falciparum cGMP-dependent protein kinase (PfPKG).[15] Although such polypharmacology is an attractive attribute in some respects, finding the balance between potency and on-target activity can be challenging in target-guided drug discovery programmes for dual inhibitors. AZD0156 was found not to inhibit PfPKG (IC50 > 10 µM, Figure 3d) in contrast to the known PfPI4Kβ/PfPKG inhibitor, sapanisertib. A subset of naphthyridine-based PfPI4Kβ inhibitors may also target the haem detoxification pathway in ABS parasites.[29] However, unlike that series, AZD0156 was found not to inhibit β-haematin (synthetic haemozoin) formation using an extracellular detergent-mediated indicator assay, relative to the standard β-haematin and haemozoin formation inhibitor chloroquine, but rather tracked with the negative control, pyrimethamine (Figure 3e).
Although AZD0165 may not necessarily have direct off-target effects on the human and Plasmodium orthologues as presented here, future studies would have to include off-target evaluations against other kinases, such as HsATM. Fortunately, the relatively lower dose coupled with a shorter length of treatment required to deal with an acute malaria infection, compared to those required for cancer, could potentially alleviate some off-target concerns.
AZD0156 Treatment Associates With PI4Kβ-Targeting Phenotypes and Killing Kinetics
An established metabolomics approach[30] was used to assess the impact of AZD0156 on parasite biochemical or metabolic pathways relative to other known Plasmodium kinase inhibitors. Drug-sensitive Pf3D7 trophozoites were treated for 2.5 h at 10 × IC50 with AZD0156, MMV390048 and sapanisertib, and the control compound atovaquone (a cytochrome bc1 complex inhibitor). The metabolic response to each compound following treatment exposure was determined by liquid chromatography-coupled mass spectrometry. Metabolite changes compared to the untreated control revealed drug-induced disruptions to key metabolic pathways. The metabolic fingerprint of AZD0156 was distinct from the control, atovaquone. The AZD0156 metaprint is more similar to that of known PfPI4Kβ inhibitor MMV390048 but less so to sapanisertib as a PfPI4Kβ/PfPKG dual inhibitor, again confirming the observed differentiation of AZD0156 from these other inhibitors and supporting its more selective PfPI4Kβ inhibition (Figure 4a).[15] Illustrated another way, principal component analysis and a heatmap correlation matrix (Figure S2), based on metabolic shifts, show the clustering of AZD0156 with other known PfPI4Kβ inhibitors rather than with compounds known to inhibit other targets. This metabolic phenotype associated with PfPI4Kβ inhibitors results in a decrease in haemoglobin-derived peptides and, while not inhibiting haemozoin formation as implied in Figure 3e above, suggests some disruption of haemoglobin catabolism via other pathways, although it should be noted that this effect is often observed for antiplasmodial agents of other classes if the test compound concentration is high enough.[30]
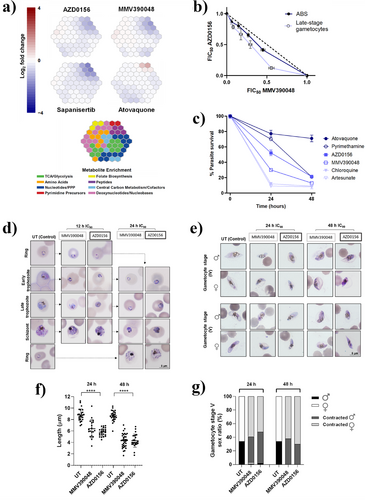
AZD0156 also behaved similarly to MMV390048 in fixed-ratio isobologram analysis on the two main life cycle stages of P. falciparum. AZD0156 combined with MMV390048 showed additive/indifferent interactions for both ABS parasites (ΣFIC50 0.9) and late-stage gametocytes (ΣFIC50 0.8) (Figure 4b). Additionally, the two compounds reacted similarly when combined with dihydroartemisinin (DHA) which acts through a nonkinase-related mechanism. When AZD0156 treatment was combined with DHA (Figure S3A), or MMV390048 together with DHA (Figure S3B), the combination was, in each instance, antagonistic for ABS parasites (ΣFIC50 1.5 for both combinations) and synergistic on late-stage gametocytes (ΣFIC50 0.9 for AZD0156 with DHA and ΣFIC50 0.5 for MMV390048 with DHA).[31]
AZD0156 showed moderate killing kinetics in the Pf3D7 strain over 48 h in an in vitro parasite reduction ratio (PRR) assay relative to known antimalarial drugs (Figure 4c). Quantitatively, while AZD0156 exhibited a faster rate of kill compared to atovaquone and pyrimethamine, the in vitro logPRR for AZD0156 indicated a slower rate of kill of 1.3 compared to other known PfPI4Kβ inhibitors, MMV390048 (logPRR 2.7)[19, 21] and UCT943 (logPRR 2.5), again suggesting some differentiation from existing PfPI4Kβ inhibitors.[21] However, because the rate of kill depends on both the mode of action and the physicochemical properties of the compound, the killing kinetics of this chemotype could potentially change during medicinal chemistry optimization. As antimalarial agents are typically used in combination, compounds with a variety of killing rates are useful in deciding the optimal combination regimen.
Both MMV390048 and UCT943 have been noted to present lag phases targeting late trophozoite stages and schizogony.[19] For AZD0156, this was confirmed to be the case through morphological evaluation, which showed minimal effect on ring-stage parasites with a 12 h treatment. However, a 24 h treatment completely halted trophozoite development, with treated trophozoites unable to enter schizogony (Figures 4d and S4). This rate of action of AZD0156 aligns with transcriptomic evaluations showing that PfPI4Kβ expression peaks during early trophozoite formation and schizogony,[32] suggesting a stage-specific phenotype that correlates to the moderately slow rate of action previously observed for MMV390048.[19]
Further morphological stage- and sex-specific evaluation of gametocytes treated with AZD0156 at IC90 for 24 and 48 h showed that gametocytes were evidently compromised (Figure 4e). Both stage IV and V gametocytes were significantly contracted (mean untreated length of 8.8 ± 0.2 µm versus AZD0156-treated length of 5.4 ± 0.4 µm, n ≥ 20, p < 0.0001, and one sample t-test, Figure 4f) already after 24 h treatment. Treated stage IV gametocytes could not mature to stage V over a 48 h treatment (Figure 4e). AZD0156 treatment affected both male and female gametocytes similarly, with no change in the sex ratio (Figure 4g). The “contracted” phenotype observed due to AZD0156 treatment correlates with that seen after MMV390048 treatment. These gametocytes are not viable and significantly metabolically compromised (decreased mitochondrial respiration as indicated by MitoTracker Red CMXRos, n = 89, p = 0.012, Mann–Whitney U test, Figure S5). The correlation of the effect of AZD0156 and MMV390048 phenotypic evaluation of gametocytes corroborates PfPI4Kβ as the target of AZD0156 in both ABS and gametocytes. The specificity of AZD0156 toward trophozoites and late-stage (IV–V) gametocytes coincides with the core roles of PfPI4Kβ within various developmental stages including DNA replication and cytokinesis[33] required in the ABS, as well as chromatin condensation for egress during gamete formation.[9, 34] These findings are consistent with previous studies showing that PfPI4Kβ is a highly-druggable multistage target in P. falciparum parasites.[9, 19, 35]
AZD0156 Shows Favorable Cytotoxicity and Pharmacokinetic Properties, and Moderate in Vivo Antimalarial Efficacy
As an indicator of potential cytotoxicity, AZD0156 was screened against the human HepG2 hepatic cell line at 2 µM for which 96% cell viability was obtained, suggesting a favorable cytotoxicity profile. A more detailed cytotoxicity study was conducted against the Chinese hamster ovary (CHO) cell line, for which AZD0156 treatment yielded an IC50 value of 11.7 µM. This results in favorable selectivity indices of >25-fold relative to the whole-cell ABS potencies measured against the three different strains as presented in Figure 1b. A lack of cardiotoxicity, through minimal interaction with the hERG potassium channel at low micromolar concentrations, has also previously been reported for AZD0156 (IC50 > 30 µM).[17] This finding further supports a favorable toxicity profile for this compound in the context of potential repositioning for malaria. Furthermore, AZD0156 was highly soluble at 195 µM in PBS (pH 6.5), compared to some other PI4Kβ inhibitors for which notably low aqueous solubility has been recorded.[21, 36]
When administered intravenously in mice, the blood clearance of AZD0156 was low (Table S1). This is consistent with the high in vitro metabolic stability observed for hepatic metabolism of this compound, viz. CLint, app (mL min−1 kg−1) < 11.6, < 11.6 and < 11.6 for human, rat, and mouse-derived liver microsomes, respectively. The plasma volume of distribution at steady state (Vss) was moderate, suggesting that the compound distributes and accumulates in organ tissues. This is expected for a basic compound due to partitioning into cell membranes associated with acidic phospholipids.[21] Consequently, the half-life of AZD0156 was moderate at approximately 4 h; this will require further optimization to achieve the desired half-life of >4 h for a single-dose cure.[37] Though not previously reported in mice, this value correlated with half-life measurements in rats.[26] Oral bioavailability was good, at almost 50%, which is encouraging in terms of potentially achieving a single-dose treatment and cure for malaria that aims to boost patient compliance in resource-limited regions of the world.
Finally, when AZD0156 was dosed orally at 4 × 50 mg kg−1 in the P. berghei malaria infection model, an 81% reduction in parasitaemia was achieved (for comparison, 99.9% and 99% reduction in parasitaemia were achieved for clinical control compounds chloroquine and artesunate, respectively, in the same model at 4 × 30 mg/kg[38]), indicating an important pharmacology proof-of-concept to motivate optimization of this compound as an oral drug for humans as a component of combination therapy for the treatment of malaria.
Conclusion
Developing new antimalarial compounds that target multiple stages of the human malaria parasite is critical to managing and reducing malaria cases and deaths effectively. Our findings demonstrate that the imidazoquinolinone AZD0156 (MMV1580483), a human ATM kinase inhibitor that completed Phase I clinical trials as a treatment for cancer, exhibits significant in vitro ABS and transmission-blocking activity against P. falciparum, the most virulent human malaria parasite. Resistance selection, cross-resistance, biochemical, and conditional knockdown studies identified PfPI4Kβ, a well-established and clinically-validated drug target for malaria, as the molecular target of AZD0156. These data were supported by metabolomic perturbation analysis, fixed-ratio isobolograms, and in vitro PRR assays, all of which phenotypically highlighted the similarity of AZD0156 to other known PfPI4Kβ inhibitors. Further profiling revealed a lack of effective biochemical inhibition of HsMAP4K4 and HsMINK1, both of which have been speculated to be associated with the toxicological signals associated with MMV390048, another PfPI4Kβ inhibitor. In that regard, AZD0156 does not display a polypharmacological profile associated with targeting other P. falciparum kinases or metabolic pathways previously observed with other PfPI4Kβ inhibitors. The advantage of this specificity for a single kinase target may contribute to the minimal off-target activity measured against mammalian kinases. These cleaner toxicity profiles suggest that this chemotype may hold fewer developmental risks. However, activity against the human anticancer target for which the compound was initially developed (ATM kinase), and risk for resistance development, will have to be closely monitored. In addition to its favorable cytotoxicity, physicochemical and pharmacokinetic properties, AZD0156 showed moderate in vivo efficacy in a P. berghei mouse malaria infection model. Taken together, these data demonstrate that AZD0156 is an attractive starting point for repositioning as an antiplasmodial lipid kinase inhibitor via structure-guided medicinal chemistry optimization, with the potential to yield molecules for much-needed therapeutic intervention against the scourge of malaria.
Supporting Information
The authors have provided additional information in the Supporting Information, viz. supplementary figures, experimental section, and computational methods.
Acknowledgements
Medicines for Malaria Venture (MMV) African Challenge Grant RD-17-0047 awarded to D.C. and L-M.B and Project RD-19-0001 to L-M.B. Future Leaders−African Independent Research (FLAIR) Fellowship Programme, a partnership between the African Academy of Sciences and the Royal Society funded by the UK Government's Global Challenges Research to L.B.C. The National Institute of Allergy and Infectious Diseases of the National Institutes of Health to K.J.W. (R01AI143521), D.A.F. (R01AI185559), and K.C. (R01AI152092). D.A.F. also gratefully acknowledges support from the Gates Foundation (INV-033538). The authors would like to thank Martha Muruya for technical support, and to acknowledge the Huck Institutes’ Metabolomics Core Facility (RRID:SCR_023864) for use of the Thermo Exactive Plus Orbitrap. We thank Ursula Lehmann for technical assistance with the P. berghei-infected mouse malaria model performed at Swiss TPH. The following reagent was obtained through BEI Resources: P. falciparum strain 3D7A (MRA-151) and P. falciparum strain Dd2 (MRA-150) contributed by David Walliker. South African Medical Research Council with funds received from the Department of Science and Innovation (SAMRC-GIPD) Drug Discovery for Malaria Elimination. The Department of Science and Innovation and the National Research Foundation South African Research Chair (SARChI) in Sustainable Malaria Control to L-M.B. (UID 84627). K.C. is the Neville Isdell Chair in African-centric Drug Discovery and Development and thanks Neville Isdell for generously funding the Chair.
Conflict of Interests
At the time of conducting this project and writing the manuscript, authors V.F. and J.G. were GSK employees and report ownership of GSK shares. No specified compensation was given to the authors in response to the development of this article. The remaining authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.