Stabilization and Binding of [V4O12]4− and Unprecedented [V20O54(NO3)]n− to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug VIVO(acetylacetonato)2
Graphical Abstract
The promising anti-diabetic and anti-cancer drug VIVO(acetylacetonato)2 in the presence of lysozyme undergoes ligand exchange, partial or complete oxidation of VIV to VV to give two polyoxidovanadates (POVs): [V4O12]4− and the unprecedented [V20O54(NO3)]n−, based on the unusual [V18O46(NO3)]n− cage. They are stabilized by non-covalent or covalent interactions with the protein.
Abstract
High-resolution crystal structures of lysozyme in the presence of the potential drug VIVO(acetylacetonato)2 under two different experimental conditions have been solved. The crystallographic study reveals the loss of the ligands, the oxidation of VIV to VV and the subsequent formation of adducts of the protein with two different polyoxidovanadates: [V4O12]4−, which interacts with lysozyme non-covalently, and the unprecedented [V20O54(NO3)]n−, which is covalenty bound to the side chain of an aspartate residue of symmetry related molecules.
Polyoxidometalates (POMs) such as coordination cages and clusters are increasingly applied in a wide range of traditional and interdisciplinary fields.1-11 These compounds exhibit interesting features in terms of molecular composition, size, solubility, shape, charge density and redox potential. Polyoxidovanadates (POVs) constitute a sub-class of the vast POMs family with remarkable physico-chemical, electronic, magnetic and—particularly—medical properties.12, 13 Although the understanding of the POVs binding to proteins is a crucial step for elucidating the mechanism of biological action of these compounds, this research field, due to its high complexity, has been scarcely investigated.14, 15 In our continuous efforts to characterize the interaction of proteins with metal complexes,16-19 we have recently studied the structures of the adducts formed upon reaction of proteins with VIVOL2 compounds, where L− is a monoanionic bidentate organic ligand.20-22 Crystallography works reporting the structures of these adducts are rare20-24 and the few studies carried out on these systems revealed binding preference of V containing fragments, in particular VIVOL+ and VIVOL2, for Asn, Asp, Glu and His side chains.
Here, we report Electron Paramagnetic Resonance (EPR) experiments and high resolution X-ray structures obtained upon exposure of hen egg white lysozyme crystals, grown under two different experimental conditions, to the complex VIVO(acac)2, where acac is acetylacetonato. VIVO(acac)2 is a water-soluble compound with therapeutic properties,25-28 that has attracted much interest for its potential use as anticancer and insulin-enhancer agent.27-29 Lysozyme is a glycoside hydrolase,30 which has been very frequently used as a model protein for metalation studies.16-19
EPR spectra were recorded as a function of time at 298 K, under the experimental conditions used to grow crystals of lysozyme treated with VIVO(acac)2. At pH 7.5, using a solution with the same composition of that from which a crystal of lysozyme in the presence of VIVO(acac)2 was isolated (structure A), spectra show that the eight isotropic resonances of VIVO (I=7/2) disappear after 4 days, suggesting the (almost) complete oxidation to an EPR-silent VV species in the timescale of crystallization experiments (Figure 1).
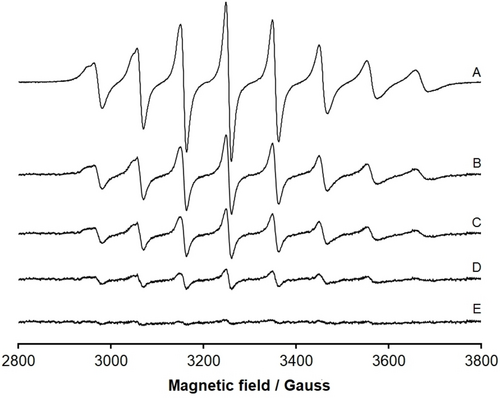
EPR spectra recorded as a function of time at 298 K on a solution containing: VIVO(acac)2 5 mM, lysozyme 20 mg mL−1, sodium formate 2 M, Hepes buffer 0.1 M, pH 7.5. (A) t=0 h; (B) t=24 h; (C) t=48 h; (D) t=72 h; (E) t=96 h. The spectra are shown using the same instrumental gain. The almost complete disappearance of the eight isotropic resonances after 4 days indicates the oxidation of VIV (3d1, EPR-active) to VV (3d0, EPR-silent).
In contrast, when the EPR spectra were measured at pH 4.0, namely in the other condition used to grow crystals of lysozyme in the presence of VIVO(acac)2 reported in this work (structure B), the reduction of the spectral intensity is evident, but—even after 96 h—the VIVO signals continue being revealed implying that the oxidation to VV is only partial (Figure S1 of Supporting Info, SI). Additional EPR experiments were carried out at pH 7.5 and 4.0 allowing O2 to flow into the solution or completely removing molecular oxygen with Ar. The results indicate that with dioxygen the oxidation is very fast and is almost complete after 24 h at neutral pH (Figures S2 and S3); in contrast, the absence of O2 slows down significantly the oxidation and the decrease of the EPR signal is negligible at pH 7.5 and not detectable at pH 4.0 (Figures S4 and S5). Overall, these data suggest that the molecular oxygen could be the oxidant in our systems.
Crystals of lysozyme were grown in: a) 2 M sodium formate and 0.1 M Hepes pH 7.5 (structure A) and b) 20 % ethylene glycol, 0.1 M sodium acetate buffer pH 4.0, 0.6 M sodium nitrate (structure B), and then were exposed to stabilizing solutions containing the mother liquors and a saturated solution of VIVO(acac)2. X-ray diffraction data were collected on these crystals at 1.18 and 1.79 Å resolution on Beamline XRD2 at Elettra synchrotron, Trieste, Italy (see Supporting Information for further details). Both crystals are isomorphous and belong to the space group P43212, with one protein molecule in the asymmetric unit. The structures refine to R-factors and Rfree values of 0.172/0.196 and 0.183/0.232, respectively (Table S1). The overall conformation of the protein is not significantly affected by the presence of the POVs. The Cα root mean square deviations from the metal-free protein structure (PDB code 193L31) are within the range 0.19–0.23 Å.
Remarkably and unexpectedly, inspection of the difference Fourier electron-density (2Fo−Fc and Fo−Fc) and anomalous difference electron density maps reveals the presence not of VIVO(acac)+/VIVO(acac)2 moieties but of POVs close to the protein surface in both the structures (Figure 2). Notably, structures A and B have been reproduced collecting X-ray diffraction data using replicates of POV/lysozyme crystals.
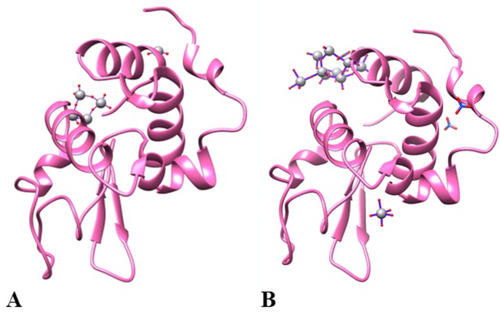
(A) Asymmetric unit content of the structures of lysozyme in the presence of VIVO(acac)2 in 2 M sodium formate and 0.1 M Hepes buffer pH 7.5 (structure A) and (B) in 20 % ethylene glycol, 0.1 M sodium acetate buffer pH 4.0, 0.6 M sodium nitrate (structure B). [V4O12]4− and a V close to two oxygen atoms are bound on protein surface in the structure A, while a [V20O54(NO3)]n− ion with a covalent binding to the side chain of Asp87 has been found in the structure B, together with an additional metal moiety with VO5 composition coordinated to the side chain of Asp52. Coordinates and structure factors of the adduct were deposited in the Protein Data Bank under the accession codes 7ZU6 and 8PTE.
In the structure A, refined at 1.18 Å resolution to R-factor/Rfree values of 0.172/0.196, non-covalent binding of [V4O12]4− on the protein surface has been found (Figure 2A). Anisotropic refinement of the structure gives R-factor/Rfree values of 0.132/0.167.
The presence of this V species suggests that the metal has changed its oxidation state from +IV to +V under the investigated experimental conditions, confirming the EPR results. An additional V atom close to two oxygens has been also found (Figure S6). [V4O12]4− anion has a cyclic structure with VV in a tetrahedral geometry and is stable in neutral solutions. [V4O12]4− is held in its position by hydrogen bonds (H-bonds) formed with the N-terminal amine, OH group of Ser86, N atom of Val2, side chain of Gln41, side chain of Asn65 from a symmetry related molecule, and water molecules (Figure 3).
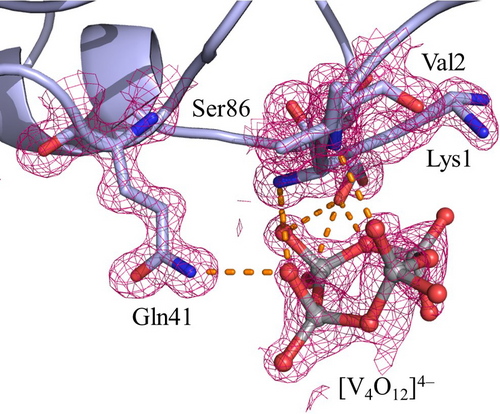
[V4O12]4− binding site in the structure A. The side chains of Lys1 and Ser86 adopt two different conformations. 2Fo−Fc electron density maps are reported at 1.0 σ level in magenta. Anomalous difference electron density is reported in Figure S7.
Binding of cyclic tetrameric oxidovanadate [V4O12]4− to proteins was observed upon self-assembling of orthovanadate in the presence of E. coli BtuCD, an ABC transporter,32 and of C3 exoenzyme from Clostridium botulinum.33 In those structures, [V4O12]4− forms H-bonds with backbone atoms and with the side chains of Asn and Thr residues.
In the structure B, obtained in 20 % ethylene glycol, 0.1 M sodium acetate buffer pH 4.0, 0.6 M sodium nitrate and refined at 1.79 Å resolution to R-factor/Rfree values of 0.183/0.232, a moiety with VO5 composition close to the side chain of Asp52 (Figure S9) and a [V20O54(NO3)]n− anion were found (Figures 2B and 4). As suggested by EPR, this species could be a POV with the simultaneous presence of VV and VIV centres. This anion can be described as an unusual octadecavanadate (V18) cage of composition [V18O46(NO3)]n− attached to two additional V atoms, each bound to five oxygens in a VO5 group where one O atom is shared with the cage; this latter group is in its turn coordinated to the side chain of Asp87 and Asp87* (i.e., the residue from a symmetry related molecule) (Figure 4). This POV is reconstituted by its symmetry mate and the occupancy is equal to 0.60. The [V20O54(NO3)]n− anion also forms a series of H-bonds with the O atoms of Glu7, Ala10, Ala11 and Ser86, the N atoms of Ala11 and Ile88 and the side chains of Lys1, Glu7, Arg14, His15, Asp87 and Thr89 (Figure 4). It is worth of being highlighted that this is the first time that a protein adduct with a V20 arrangement, and in general with a POV greater than V10, is reported in the literature and the first time that a covalent binding of a protein to a POV is observed.14 Moreover, a search in the Cambridge Structural Database (CSD) reveals that there are not known structures of POVs with 20 V atoms. Instead, in literature, five types of octadecavanadate (V18) cages with composition [V18O39]n−,34 [V18O41]n−,35 [V18O42]n−,36-45 [V18O44]n−,46, 47 and [V18O46]n−,48 have been reported. These present various combinations of VIV and VV centres,36 from which the value of the charge n−, the structural features and the magnetic,37 catalytic,49, 50 and biological properties depend.46, 51 These POVs are able to incorporate in the centre of the cage various types of anions such as Cl−, Br−, I−, HS−, HCOO−, CO32−, N3−, NO2−, NO3− or, also, neutral species like water. To the best of our knowledge, the cage observed in our structure is the second one with composition V18O46, after [VV16VIV2O46(NO3)]5−.48 Notably, even though the V18 cage observed in the structure B resembles that with V18 composition [VV16VIV2O46(NO3)]5− (reported in the CSD under the accession code UFIQAC48), a comparison between the two structures reveals significant differences (Figure 5). In fact, while the anion [VV16VIV2O46(NO3)]5− has all V atoms with square pyramidal geometry based on VO5 group, the cage found in the structure B has two V atoms with tetrahedral geometry (like the VO4 group) (Figure 5); notably, the V centers of VO5 groups, that lead to [V20O54(NO3)]n− anion and covalently interact with Asp87 and Asp87* residues, are bound to these tetrahedral vanadium atoms. So, the possibility of a different combination of VIV and VV centres in the two structures cannot be excluded nor the influence of protein in the stabilization of V20O54 POV found in this study.
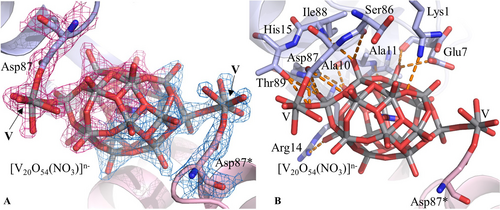
[V20O54(NO3)]n− binding site in the structure B (panel A) and its interaction with protein residues (panel B). 2Fo−Fc electron density map is reported at 1.0 σ level in red (cyan for the symmetry mate). Asterisk (*) indicates atoms from symmetry related molecules. Anomalous difference electron density map is reported in Figure S8.
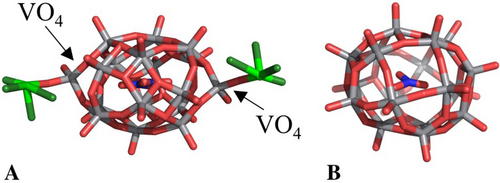
[V18O46(NO3)]n− cage found in the structure B of this work (panel A) and [VV16VIV2O46(NO3)]5− anion reported in ref. [48] (panel B). In panel A the two V atoms that are bound covalently to the protein (reconstituting the [V20O54(NO3)]n− anion) and their oxygen ligands are reported in green.
Overall, these data highlight the potential stabilizing role of proteins toward unusual or transient POVs either by the direct coordinative bond or by H-bonds and van der Waals interactions.
In conclusion, here we have reported two structures of lysozyme with POVs which deserve three different types of comments.
-
Formation of POVs with protein crystals was already observed in a few previous studies,12-14 but V oxide polyhedra that were identified up to now were limited to [V2O7]4−, [V3O10]5−, [V4O12]4−, [V4O13]6−, [V7O19]3− (although some doubts about the interpretation of this latter structure have recently been advanced14), and [V10O28]6−. Our structural data enlarge the repertoire of structures of known protein-POVs adducts and provide useful information on the recognition of POVs by proteins. In particular, we have reported an unusual example of protein structure in the presence of the unprecedented V oxide polyhedron [V20O54(NO3)]n−. Furthermore, a new arrangement of the [V18O46(NO3)]n− cage is reported for the first time, as well as their interaction with a protein.
-
Our results show once again that protein crystals can serve as chemical reaction vessels and that soaking of metal compounds within protein crystals coupled to the determination of the X-ray structure is a strategy to capture unexpected metal compound structures, like POVs formed from solutions containing different V species. Data also suggest that protein crystals can be used to stabilize elusive POVs, providing nice examples of stabilization of unrevealed POV structures by formation of electrostatic interactions and H-bonds within the protein crystal packing.
-
From a biological point of view, the results indicate that VIVO(acac)2 undergoes dissociation, loss of the ligands, vanadium oxidation, and subsequent reassembly, forming POVs with a different number of V atoms ([V4O12]4− and [V20O54(NO3)]n−), depending on the used experimental conditions such as solvent, pH, concentration of the V compound, etc., which—in their turn—can interact with the protein. The consequence is that, under physiological conditions, biologically VIVOL2 active species formed by L− ligands with weak or intermediate strength could give, at least partially, one or more POVs which—in turn—can interact with proteins and biomolecules forming adducts that could contribute to the transport in organism and pharmacological activity of the potential drugs.
Our results provide new structural basis for interpreting experiments carried out with the potential drug VIVO(acac)2 and similar VIVO-based drugs and for understanding their significant biological properties. Indeed, as observed in the literature, to throw light on the mechanism of action of these compounds, their speciation in biological fluids and in the cell milieu and possible subsequent oxidation reactions should be studied in detail.52, 53 Here, in particular, we have demonstrated that VIVO(acac)2 can degrade, oxidize, and form vanadium(V) oxide polyhedra which—in turn—can interact with the proteins forming covalent or non-covalent adducts stabilized mainly by H-bonds with backbone atoms and side chains of Arg, Asn, Asp, Glu, His, Lys, and Thr residues.
Acknowledgments
The authors thank Elettra staff for their assistance during data collection. This research is funded by the grant Fondazione di Sardegna - 2017 (FdS2017Garribba).
Conflict of interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.





