Site-Specific Ubiquitination of Tau Amyloids Promoted by the E3 Ligase CHIP
The earlier version contained the wrong Supplementary version. The Supplementary Information has been replaced with the correct version
Graphical Abstract
Abstract
Post-translational modifications of Tau are emerging as key players in determining the onset and progression of different tauopathies such as Alzheimer's disease, and are recognized to mediate the structural diversity of the disease-specific Tau amyloids. Here we show that the E3 ligase CHIP catalyzes the site-specific ubiquitination of Tau filaments both in vitro and in cellular models, proving that also Tau amyloid aggregates are direct substrate of PTMs. Transmission electron microscopy and mass spectrometry analysis on ubiquitin-modified Tau amyloids revealed that the conformation of the filaments restricts CHIP-mediated ubiquitination to specific positions of the repeat domain, while only minor alterations in the structure of the fibril core were inferred using seeding experiments in vitro and in a cell-based tauopathy model. Overexpression of CHIP significantly increased the ubiquitination of exogenous PHF, proving that the ligase can interact and modify Tau aggregates also in a complex cellular environment.
Introduction
The microtubule-associated protein Tau is an intrinsically disordered protein (IDP) highly expressed in neurons, where it participates in the assembly and stabilization of microtubules.1, 2 The abnormal accumulation of insoluble fibrillar aggregates of hyperphosphorylated Tau protein in neurons, referred as neurofibrillary tangles, characterizes a wide and heterogeneous class of neurodegenerative disorders collectively referred as tauopathies.3, 4 These diseases present a broad range of clinical phenotypes and are traditionally divided into primary tauopathies, such as corticobasal degeneration (CBD) and Pick's disease (PiD), and secondary tauopathies, primarily Alzheimer's disease (AD), which are characterized by the concomitant presence of intracellular neurofibrillary tangles and extracellular amyloid plaques.5-7 The recent advances in structural characterization of amyloid filaments obtained from pathological brain tissues8-12 have provided insights at atomic level into the different conformations that misfolded Tau can adopt in the fibril core.13, 14 The type and extent of post-translational modifications (PTMs) on Tau are emerging as major players in directing the formation of the disease-specific strain.15-17 Although the abnormal hyperphosphorylation of Tau is considered the initial step in promoting its dissociation from microtubules and aggregation, a series of different PTMs, including mono and polyubiquitination, have been identified in the filaments at the late stages of pathology.18 If on one hand the accumulation of ubiquitinated Tau suggests the failure of proteasome-dependent or autophagosome clearance of toxic species in damaged neurons, the identification of ubiquitin-modified residues in paired helical filaments (PHF) suggests a possible structural role of this bulky modifier in the formation and stabilization of Tau aggregates.15
CHIP (carboxyl terminus of Hsp70 interacting protein) is a RING/U-box type E3 ubiquitin ligase that promotes the ubiquitination of client proteins bound to Hsc70/Hsp70 or Hsp90 chaperones, directing their proteasome-mediated degradation and thus preventing the accumulation of misfolded proteins in the cell.19-21 It has been demonstrated that CHIP directly interacts with Tau and promotes its ubiquitination both in vivo and in vitro.22-24 The co-localization of CHIP with tau lesions, including neurofibrillary tangles, in postmortem brains of several tauopathies suggests a possible involvement of CHIP-mediated ubiquitination of tau fibrils in pathological events.23 Moreover, the accumulation of insoluble Tau aggregates in central nervous system (CNS) of CHIP-/- animal models reveals its essential role in the ubiquitin-dependent clearance of phosphorylated Tau isoforms in vivo.25 In our previous work we studied the structural determinants underlying the molecular recognition and interaction between Tau and CHIP in solution,26 and we reported that CHIP is able to ubiquitinate the isolated four-repeat domain of monomeric Tau (Tau4RD, also known as K18 fragment) at multiple sites in vitro.27 We have shown that the mixture of monoubiquitinated tau isomers and specific ubiquitination at Lys311 and Lys317 inhibited the formation of filaments.28 The observation that pathological tau aggregates isolated from postmortem AD brains are decorated with ubiquitin at Lys 311 and 317 led us to speculate that ubiquitination at these sites may originate from a modification event that follows the formation of the fibrils core. Despite all evidence, it remains unclear whether CHIP can directly target Tau molecules also when they are part of insoluble fibrillar aggregates.
Here, we present new results on the direct ubiquitination of different Tau filaments by CHIP, proving the ability of the ligase to target preformed Tau fibrils in vitro and within living cells. We also describe the site specificity of the ligase for different filament conformations, including pathological AD PHF, and characterize the structural properties of the modified aggregates.
Results and Discussion
CHIP directly interacts with Tau filaments and promotes their ubiquitination
It is known that the E3 ligase CHIP interacts in vivo with soluble Tau and catalyzes its ubiquitination also in the absence of Hsp70/Hsc70 chaperones23. We hypothesized that CHIP might interact with Tau filaments and direct their ubiquitination as it does with the monomeric soluble protein.27, 26 Since most of the ubiquitin-modified residues in AD brain filaments are included in the repeat domain (RD),18 we initially focused on Tau4RD preformed filaments (PFF) (Figure S1A). Tau4RD construct aggregates only in the presence of polyanionic factors such as heparin29 and it has been extensively characterized,30 therefore representing a good model for our investigation. Heparin-induced PFF, purified to remove any trace of soluble monomer (Figure S1B), were used in co-sedimentation assays with his-tagged CHIP. After incubation with PFF and repeated washing, a substantial part of the input protein was found in the insoluble fibril fraction, indicating a rather strong binding affinity of CHIP towards PFF (Figure S1C). Heparin-induced Tau filaments, however, are highly polymorphic and structurally different from those observed in AD brain.30, 31 It has been reported that a shorter fragment spanning residues 297–391 of Tau (corresponding to the proteolytically stable core of AD PHF, also known as “dGAE”) is able to form filaments structurally identical to those found in AD brain in absence of polyanionic cofactors,32, 33 and a detailed method to reconstitute homogeneous AD PHF samples in vitro has been recently published.34 So far, this is the only recombinant tau fragment found capable of reproducing the pathological conformation of AD in vitro. Following the described protocol, we prepared Tau297–391 PHF and demonstrated that they also directly bind to the ligase (Figure S1D–E). When the same experiments were performed with ubiquitin (Figure S1 C and E, lanes 5–8) or bovine serum albumin (BSA) as input (Figure S1F), virtually no protein was found bound to the insoluble fibril fraction, suggesting that CHIP binding to Tau assemblies is specific and not due to a generic protein-fibril interaction.
Next, we assessed the ability of CHIP to catalyze the ubiquitination of the preformed Tau filaments. In our previous work, we used CHIP, in combination with UBE2N (but in absence of UBE2V1 or UBE2V2), to direct monoubiquitination on monomeric Tau4RD in vitro.27 Here, we employed a modified protocol in which the substrate consisted of purified Tau fibrils instead of the monomer. The experimental workflow is outlined in Figure 1A. After 5 hours of reaction, Tau filaments isolated by centrifugation were solubilized and analyzed by SDS-PAGE. The presence of a new band at ≈25 or 19 kDa, consistent with the size of monoubiquitinated Tau4RD or Tau297–391 respectively, indicated that Tau assemblies were successfully modified under these conditions. These results were further confirmed by immunoblotting with anti-ubiquitin and anti-Tau antibodies (Figure 1B). Densitometric analysis of the gel bands from three independent reactions revealed a rather constant degree of ubiquitination (17.3±0.6 % of Tau4RD and 14±1.1 % of Tau297–391, respectively).
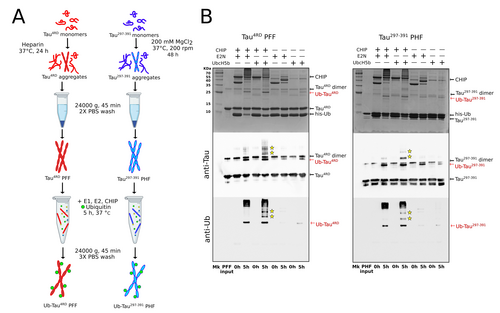
CHIP promotes ubiquitination of preformed Tau filaments. A) Schematic diagram showing the main steps of Tau filaments ubiquitination in vitro. B) Upper panels: SDS PAGE stained with Coomassie blue illustrating the ubiquitination reaction products at t0 and after 5 h of incubation of Tau4RD PFF (left) and Tau297–391 PHF (right). The same samples were analyzed by immunoblotting with anti-Tau (middle panels) and anti-mono- and polyubiquitinated proteins antibodies (lower panels). Polyubiquitinated Tau species are indicated with yellow stars. Uncropped gels and blots are shown in Figure S12.
It is known that CHIP, in addition to UBE2N, can function with a number of E2 enzymes.35 Among them, UbcH5b (also known as UBE2D2) has been shown to promote CHIP-mediated polyubiquitination of Tau protein in vitro,23 and it has been proposed as a physiological partner of CHIP in hyperphosphorylated Tau clearance in vivo.36 When UbcH5b is used as the E2 enzyme in the reaction, in addition to monoubiquitinated Tau, minor amounts of high molecular weight species are detected with both antibodies, suggesting the formation of polyubiquitinated species in both types of filaments (Figure 1B). In absence of CHIP, however, only traces of modified Tau are found, indicating that the E3 ligase is essential for the recognition and modification of Tau assemblies.
We then used transmission electron microscopy (TEM) to investigate the effect of ubiquitination on the morphology of the filaments (Figure 2, Figure S2–S3). In the case of heparin-induced Tau4RD filaments, a mixed population of twisted and untwisted filaments was observed, without any significant difference in the overall shape of PFF upon ubiquitination. Similarly, no macroscopic alteration is clearly visible in ubiquitinated Tau297–391 PHF. The morphological analysis (Figure 2C–D, Table S2) revealed no statistically significant variations in the average narrow and large diameter and crossover repeat distance, with values in line with what previously reported for Tau4RD filaments37 and recombinant Tau297–391 PHF.33 Together, the data indicate that the modification of some lysine side chains, even with a bulky ubiquitin molecule, is not sufficient to destabilize the assembly nor to produce appreciable alterations in the filament core.
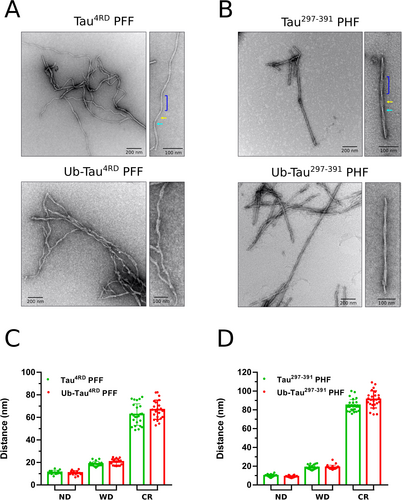
Ubiquitination does not modify filament morphology. Negative stain TEM images of unmodified and ubiquitinated Tau4RD PFF (A) and Tau297–391 PHF (B). An enlarged view of a single filament is shown on the right. The narrow (cyan arrow) and wide (yellow arrow) sections and the crossover repeat distance (blue bracket) are indicated in the top-right panels. C–D) Morphological analysis of Tau4RD and Tau297–391 filaments. The histograms display the average±SD of narrow (ND) and wide (WD) diameter and crossover repeat distance (CR) of unmodified (red) and ubiquitinated (green) filaments. Measurements taken at different points along individual fibrils, collected from at least 3 different micrographs, are indicated by green and red dots. Statistics are reported in Table S2. Student's t-test between unmodified and ubiquitinated filaments revealed no significant differences in any of the values for both Tau4RD and Tau297–391.
We then performed immunogold labelling, with anti-Ub or anti-6XHis (targeting the N-terminal His-tag of ubiquitin) primary antibodies, of the ubiquitinated filaments Tau4RD PFF and Tau297–391 PHF, to detect and localize the covalently bound ubiquitin moieties on the fibril surface (Figure 3, S4–S6). In both cases, gold particles are clearly visible, albeit to different extents, on the surface of most of the imaged filaments, indicating the presence of ubiquitin-modified protomers in the Tau assemblies.
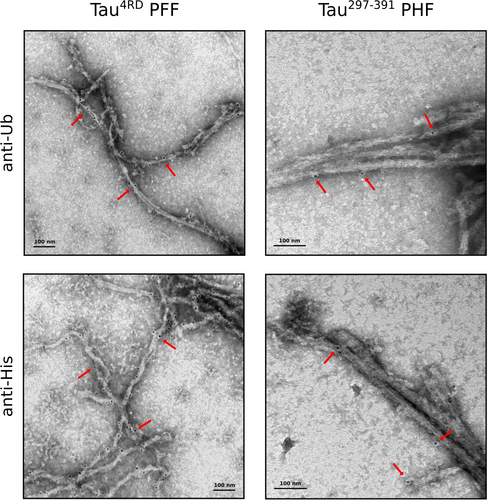
Localization of ubiquitin-modified sites on Tau filaments. Immunogold labelling of ubiquitinated Tau4RD PFF (left) and Tau297–391 PHF (right) samples with anti-Ub or anti-6XHis primary antibody and secondary antibody conjugated to 5 nm gold particles (black dots). Examples of immunoreactive regions on the surface of the filaments are indicated by the red arrows.
In general, heparin-induced filaments showed better labeling, with no clear preference for twisted or flat ribbon-like structures. Ubiquitinated Tau297–391 samples, on the other hand, suffered more from the immunogold staining process and appeared as morphologically less defined bundles of aggregated filaments with fewer bound gold particles. In both cases, however, the lack of any clear pattern in the position of bound gold particles suggests a reaction mechanism in which the ligase transiently interacts and modifies lysine residues in random positions along the filament axis regardless the local morphology.
The filament structure restricts ubiquitination to specific regions of Tau amyloids
In previous work, we characterized the product of the enzymatic ubiquitination of monomeric Tau4RD by CHIP.26 A total of 12 lysine residues were found to be targeted by CHIP (Table S3), most of which were also found in a comprehensive quantitative proteomics analysis of Tau filaments from AD patients (Figure 4A).18 We therefore set out to investigate which of these residues remained accessible for modification also in preformed filaments. Mass spectrometry analysis revealed that modification took place at only 7 out of the 12 positions (Figure 4B, Table S4, SupportingDataS1). It is worth noting that most of the residues modified in repeats R1-R2 and R4 in the monomer were found ubiquitinated in PFF as well, while almost all sites (the only exception being lysine 311, although with very low occurrence) in the sequence spanning the end of R2 and all R3 resulted unmodified in the filaments. Furthermore, the highest occurrence of ubiquitination in PFF was observed in two positions of R4 (353 and 369/370), whereas K267, K298 and K353 were the most frequently modified in Tau4RD monomers (Figure S7A).
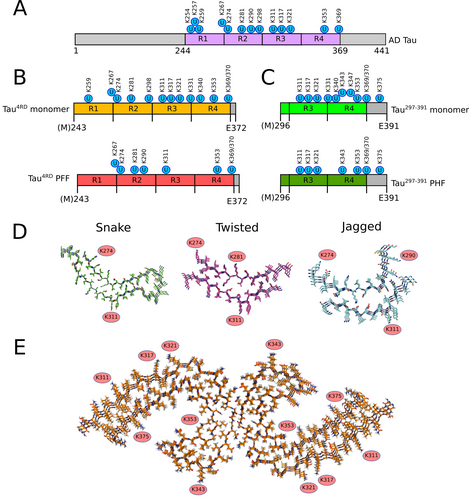
Filament core structure restricts ubiquitination at specific positions. Schematic diagrams displaying the ubiquitinated residues identified in Tau filaments from AD patients (A),18 the sites ubiquitinated by CHIP in Tau4RD (B)27 and Tau297–391 (C) monomers (upper) and filaments (lower). D) Accessible lysine side chains ubiquitinated by CHIP in snake, twisted and jagged Tau4RD filament models (PDB codes: 6QJH, 6QJM and 6QJP30). E) Position of lysine side chains ubiquitinated by CHIP in Tau297–391 PHF model (PDB code: 7QL4).
Recently, the cryoEM structures of heparin-induced filaments of recombinant 2N4R Tau have been reported.30 At least four different types of filaments have been observed, three of which at atomic resolution. These alternative conformations, named “snake”, “twisted” and “jagged”, share a similar kinked hairpin fold with β-strands spanning mainly R2 and R3, and are remarkably different from the structure of filaments from Alzheimer's and Pick's diseases, where β-structures cover all the repeat domain and extend beyond R4.8-10 We therefore mapped the position of the ubiquitinated residues in our Tau4RD PFF on these available atomic models (Figure 4D). While K274 and K311 are solvent-exposed in all the structures, K281 is in outward-facing position and thus accessible for ubiquitination only in twisted filaments, while its modification is very unlikely in the other two conformations. Similarly, K290 appears to be rather accessible only in jagged filaments. Interestingly, two spatially close residues, K317 and K321, solvent-exposed in all types of filaments, are ubiquitinated by CHIP in soluble Tau4RD monomer but not in PFF, as happens for K331 (which however is not visible in cryoEM structures). In the structures, these side chains face an extra density, associated with negatively charged groups of heparin, that mask the surface of the filament30 and that may explain why they are not accessible for modification.
Next, we analyzed the site specificity of CHIP with the pathological conformation of AD (Figure 4C). The soluble monomeric peptide Tau297–391 was found to be ubiquitinated at 10 of its 14 lysine residues, with the highest occurrence at positions K321, K347, K353 and K375 (Table S5, SupportingDataS2). In contrast, when assembled into PHF, it was modified in only 7 of these positions (Table S6, SupportingDataS3), with a similar occurrence rate as the monomer (Figure S7B). No ubiquitination was observed in PHF at positions 331 and 340, which are located at the interface between the two protofilaments and thus are probably inaccessible (Figure 4E). In contrast to heparin-induced filaments, solvent-exposed side chains of K317 and K321 was found modified in Tau297–391 PHF, the latter with remarkably higher occurrence than the monomer. Interestingly, these positions were found ubiquitinated in Tau PHF extracted from brains of AD patients, with increasing frequency with disease progression,18 and additional densities compatible with the presence of a (poly)-ubiquitin moiety has been observed in cryoEM structures of AD brain PHF.8, 15 Unfortunately, nothing can be deduced about the lysine residues included in R1 and R2, as no bona fide PHF conformation with longer fragments has been obtained in vitro so far.
Taken together, these results suggest the importance of the specific cross-β structures adopted by the filament core in limiting the interaction with E3 ligase to more localized portions of the Tau assemblies and in determining the site specificity of the modification.
Ubiquitination does not impair filament seeding activity
The self-assembly of Tau into insoluble filaments proceeds through a well characterized nucleation-elongation mechanism whereby the formation of oligomeric species in the initial lag phase is followed by the fast increase of fibril size through the recruitment of monomeric units.38 The nucleation process can be enhanced by the addition of small amounts of preformed fibril seeds, and it has in fact been shown that Tau4RD PFF promote the aggregation of different Tau isoforms both in vitro39, 40 and in cellular models.41, 42 Therefore, we sought to assess whether ubiquitination might alter, to some extent, the seeding properties of Tau filaments. First, the effect of modified Tau4RD PFF seeds on the aggregation kinetic was evaluated in Thioflavin T fluorescence-based aggregation assays (Figure 5A). The addition of 0.5 and 1 μM sonicated PFF or Ub-PFF resulted in similar, dose-dependent increase of the aggregation rate and greater variability among the individual replicates compared to the control, as expected in the presence of seed-induced secondary nucleation processes.43, 44 We note that the rapid aggregation kinetics under these experimental conditions does not allow a reliable estimate of the lag time and therefore we used the estimated t0.5 for comparison. The similar reduction in the calculated t0.5 from 87±6 min (non-seeded) to 47±6 and 39±8 min (1 μM unmodified and ubiquitinated seeds respectively) suggested that the presence of modified residues has only a marginal impact on their seeding activity in vitro. A similar behavior was observed in aggregation experiments with Tau297–391, where the addition of native or ubiquitinated PHF seeds induced a reduction of the lag time (tlag) and an earlier onset of the exponential elongation phase compared to the unseeded control (Figure 5B). In this case, however, the insertion of preformed fibril seeds (regardless the presence of ubiquitin-modified residues) also significantly reduced the steepness of the exponential phase, indicating a more complex modulation of the aggregation kinetics.
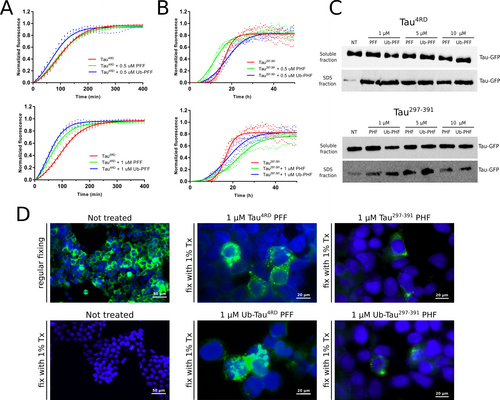
Effect of ubiquitination on Tau filament seeding activity. ThT aggregation assay of Tau4RD (A) and Tau297–391 (B) unseeded (red) and in presence of 0.5 μM (upper panels) or 1 μM (lower panels) unmodified (green) or ubiquitinated (blue) seeds. The continuous lines represent the best fit calculated on three independent replicates (dots) as described in the experimental procedures. C) western blot analysis of soluble and insoluble Tau-GFP in G12 cells treated with unmodified or ubiquitinated seeds. Soluble and insoluble cellular fractions were probed with Tau5 primary antibody. The same amount of total protein was loaded in each lane. NT: untreated control. D) Left panels: GFP fluorescence in untreated G12 cells after regular fixing (top) and with fixing in presence of 1 % Triton-X100 (bottom) to remove the soluble proteins. Middle panels: cells treated for 72 h with 1 μM Tau4RD PFF (top) or Ub-Tau4RD PFF (bottom). Right panels: cells treated for 72 h with 1 μM Tau297–391 PHF (top) or Ub-Tau297–391 PHF (bottom). Tau-GFP aggregates are clearly visible in some cells as strong localized fluorescence signals. Nuclei were stained with DAPI (blue) for better visualization. The results displayed in C and D are representative of two independent experiments. Uncropped blots of panel C are shown in Figure S12.
Previous reports have highlighted the ability of exogenous Tau4RD seeds to trigger the aggregation of soluble Tau in cellular models.45, 46 We therefore generated a Hek293 cell line stably expressing GFP-tagged full-length human tau (2N4R) carrying the aggregation-prone P301L mutation (herein referred to as Clone G12) and investigated the effect of ubiquitination on the seed-induced aggregation of cellular Tau. We transduced G12 cells with different concentrations of unmodified or ubiquitinated Tau4RD PFF and Tau297–391 PHF for 72 hours and evaluated the amount of Tau-GFP present in the soluble and insoluble fractions. At all concentrations tested (1, 5 and 10 μM), both type of seeds induced the accumulation of endogenous Tau aggregates, regardless the presence of ubiquitin-modified residues, whereas a significantly lower level was found in the untreated control (Figure 5C). Intracellular aggregates of Tau are clearly visible as focal fluorescent inclusions in cells treated with filament seeds (1 μM) when fixed in presence of 1 % Triton (Figure 5D). Overall, these findings suggest that ubiquitination does not substantially modify the propensity of Tau filaments to promote nucleation and aggregation of the soluble cytoplasmic protein.
CHIP promotes ubiquitination of internalized PHF in cells
We next investigated whether CHIP could support the ubiquitination of exogenous Tau PHF also in living cells. We first generated a CHIP knock-out cell line (Stub1−/−) in which the expression of the endogenous ligase was totally abolished (Figure S8). Then, we transduced Tau297–391 PHF into this negative background and in Stub1−/− cells overexpressing recombinant myc-tagged CHIP. PHF (but not the monomer) are efficiently internalized after 16 hours of treatment, as assessed by western blot and immunofluorescence analysis with an anti-Tau antibody (Figure S9A–B). Notably, only a slight decrease in viability was observed in cells treated for 3 days with different concentrations of PHF (Figure S9C), whereas the amount of internalized filaments remained essentially unchanged (Figure S9D–E). CHIP-mediated ubiquitination of exogenous PHF was initially studied by confocal microscopy. Stub1−/− cells, transfected with myc-CHIP or empty vector, were transduced with Tau297–391 PHF, and the Triton-insoluble fraction was stained with anti-Tau and anti-Ub antibodies (Figure 6A, Figure S10). Overexpression of CHIP resulted in the presence of large, ubiquitin-positive insoluble aggregates, some of which clearly colocalized with Tau. A pixel-matching analysis on at least 10 randomly selected fields, expressed as Pearson's correlation coefficients, revealed a statistically higher degree of colocalization of the two signals (Ub and Tau) when CHIP is expressed (Figure 6B).
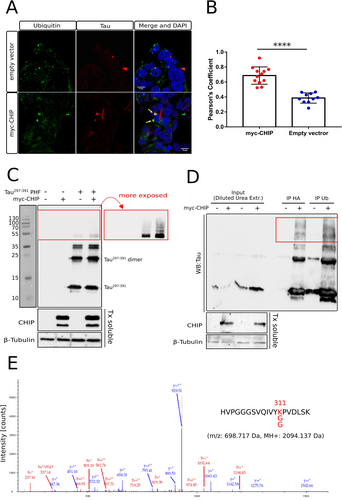
CHIP promotes the ubiquitination of internalized PHF in cells. A) Representative Z-plane images of Stub1−/− cells after 24 hours treatment with Tau297–391 PHF in presence or absence of CHIP. Cells were fixed in presence of 1 % Triton X-100 to remove the soluble proteins and stained with anti-Tau (red) and anti-Ub (green). Scale bars: 10 μm. Examples of colocalized fluorescent signal in the merged images (yellow) is indicated by arrows. B) 3D colocalization analysis for ubiquitin and PHF expressed as Pearson's correlation coefficients. Results from 12 (CHIP) and 10 (empty vector) random fields in which both signals were present, selected from two independent replicates, are shown as blue and red dots on the histograms (errors bars: ±SD) (****, p<0.001, unpaired T-test). C) western blot analysis of cellular insoluble fractions after the treatment with Tau297–391 PHF. Stub1−/− cells, transfected with HA-ubiquitin and myc-CHIP (or empty vector), were treated for 16 h with 5 μM PHF or vehicle. Triton-insoluble fractions solubilized with 1 % SDS were probed with anti-Tau antibody. A more exposed blot of the same membrane is shown in the red box. CHIP and β-tubulin (loading control) in the soluble fractions are displayed below. D) Immunoprecipitation of cellular insoluble fractions with anti-Ub or anti-HA after the treatment with Tau297–391 PHF. HMW Tau297–391 species are highlighted with a red box. CHIP and β-tubulin (loading control) in the Triton-soluble fractions are displayed below. E) Fragmentation mass spectrum of the K-GG modified Tau peptide identified in Tau297–391 PHF in cells overexpressing CHIP. m/z: 698.72 Da, Charge: +3, MH+: 2094.14 Da. The ubiquitinated position (K311) is highlighted in red in the sequence. Uncropped blots of panels C and D are shown in Figure S12.
We then looked for the presence of ubiquitin-modified Tau protomers in filaments internalized by Stub1−/− cells overexpressing recombinant CHIP and HA-tagged ubiquitin. The insoluble cellular fractions, after 16 h of incubation with PHF and 5 hours of treatment with the proteasome inhibitor MG132, were solubilized with SDS and analyzed by western blot with anti-Tau antibody. Although Tau was found to be mostly unmodified even in the presence of CHIP, longer exposures revealed greater accumulation of modified, high-molecular-weight (HMW) Tau297–391 species when the ligase was present (Figure 6C). To verify the occurrence of covalently-bound ubiquitin in such species, the insoluble fractions containing internalized PHF were solubilized in 8 M urea and, after dilution, subjected to immunoprecipitation with anti-Ub or anti-HA antibodies. Enrichment of HMW species in immunoprecipitated samples indicated a marked increase in ubiquitin-modified Tau when CHIP is overexpressed (Figure 6D). HMW bands from insoluble cellular fractions were then extracted from gel, trypsin digested and analyzed by mass spectrometry. Although the amount of Tau protein in the samples was very low, we were able to detect an ubiquitin-modified Tau peptide at position 311 in PHF from cells expressing CHIP, while the same peptide was found only unmodified in the control (Figure 6E, S11, SupportingData S4 and 5).
Overall, our results showed that CHIP can promote the ubiquitination of Tau filaments even in a complex cellular environment. Noteworthy, while CHIP-mediated modification of PHF in vitro with the E2 enzyme UBE2N predominantly resulted in monoubiquitinated Tau monomers (Figure 1B), higher molecular weight species were prevalent in cells, similarly to what observed in vitro with E2 UbcH5b.
Discussion
In the last years, advances in cryo-EM have enabled unprecedented insights into the structural heterogeneity of Tau amyloid fibrils. Following the first atomic structures of Straight and Paired Helical Filaments purified from AD brains,8, 10 the architecture of Tau amyloids from different neurodegenerative diseases have been described,9, 11, 12 prompting the introduction of a novel, structure-based classification of tauopathies.47 Although hyperphosphorylation has been widely recognized as the early event triggering the dissociation from microtubules and aggregation of Tau in the initial stages of the disease, other PTMs, and in particular ubiquitination, are emerging as key players in determining the progression and the final structure of the filaments in different tauopathy strains.15, 18
In vivo, Tau is known to be ubiquitinated only by few E3 ligases, such as axotrophin/MARCH7, TNF receptor-associated factor 6 (TRAF6) and CHIP.23, 48, 49 In our recent works, we demonstrated that CHIP modifies the isolated Tau4RD domain at multiple sites, and that ubiquitination altered the aggregation kinetics of Tau in a site-specific manner.27, 50 In particular, the presence of ubiquitinated residues within the RD in Tau aggregates isolated from AD patients,18, 51 in positions that we found to disfavor aggregation in our in vitro model, and the extent of previous biochemical and structural characterization of this domain,29 prompted us to investigate whether CHIP can target preformed Tau filaments as it does with the monomeric protein. Initially, we used purified Tau4RD PFF as reaction substrate, and we identified ubiquitin-modified residues in ≈20 % of the Tau protomers in the fibrils, although in a more restricted range of positions compared to what observed for the soluble monomeric protein. Most of the sites in R3 resulted inaccessible for modification in PFF, indicating that the cross β-structure adopted by the filament core influences the interaction with the E3 ligase and the accessibility of certain lysine side chains. However, cryo-EM studies revealed that heparin-induced Tau aggregates are highly polymorphic, with at least four types of filaments obtained with the 2N4R Tau isoform.29 More recently, the structural characterization of Tau 0N4R fibrils by solid-state NMR revealed a single β-sheet conformation for the fibril core, different from the structures determined by cryo-EM.52 It has been hypothesized that the polymorphic nature of heparin-induced filaments may largely be due to the experimental conditions, thus making it difficult to compare the results obtained by different groups. In this view, it is possible that the ubiquitination pattern we observed in our Tau4RD PFF may also be affected, to some extent, by the structural heterogeneity of the substrate. Besides being polymorphic, all Tau filaments obtained in presence of heparin or other polyanionic cofactors are structurally different from those isolated from AD and other tauopathies patients. Recently, bona fide AD PHF have been obtained in vitro using the recombinant fragment 297–391, which spans repeats 3 and 4 of Tau and an additional stretch of residues at the C-terminus.32 When these filaments were used as substrates in our ubiquitination reaction, again the most frequently modified positions correspond to solvent-exposed lysine residues in PHF structures.
Emerging evidence suggests that pathological Tau species can propagate through a “prion-like” mechanism, in which released fragments of preformed fibrils are taken from a receiving neuronal cell and seed the aggregation of the endogenous protein.41, 44, 53, 54 The cross-seeding properties of PFF are strictly related to the conformation of the β-structures in the fibril core: it is known that Tau4RD (K18) fibrils can seed homotypic 4R, but not 3R (K19), and full-length Tau monomers in vitro,55, 40 and can promote the formation of neurofibrillary tangles-like aggregates in cultured cells and neurons.42, 56 Ubiquitinated Tau filaments did not show appreciable structural changes in shape and morphology compared to the unmodified counterparts, and displayed very similar seeding activity towards Tau aggregation both in vitro and in cellular models. These observations led us to suppose that the modification of some residues on the filament surface, even with bulky ubiquitin moieties, is not sufficient to induce substantial alterations in the structured fibril core.
The accumulation of ubiquitin-modified Tau aggregates has been associated with a misregulation of their turnover and with a reduced clearance of abnormal, ubiquitinated Tau species by autophagy and proteasomal pathways.23, 51, 57 An open question is whether any ubiquitin ligase can target Tau also in the form of insoluble filaments. In fact, the role of amyloid fibrils as substrates of E3 ligases is still poorly understood, and only few examples have been described so far. Members of Nedd4 ligases family have been shown to efficiently recognize and ubiquitinate α-synuclein fibrils in vitro,58 and subsequently a cullin-RING ubiquitin ligase that promotes ubiquitination of internalized insoluble α-synuclein has been reported to facilitate the clearance of toxic amyloid species in cultured cells.59 Using a knockout cell line, we demonstrated here that CHIP overexpression can increase the ubiquitination of exogenous Tau PHF within the cell, showing its ability to target preformed amyloid aggregates also in a complex cellular environment. Indeed, in the presence of CHIP we detected an ubiquitin modification at position 311 of Tau in internalized PHF, and this result is in line with previously reported data showing that K311 serves as acceptor of ubiquitin in PHF extracted from AD brain.18, 51, 57
Conclusion
Besides its well-known activity on soluble Tau protein, we found that the E3 ligase CHIP can promote the in vitro ubiquitination of preformed Tau amyloids, with a site specificity that depends on the filament structure. In addition, we demonstrated that CHIP is also able to mediate the ubiquitination of exogenous, AD-like filaments in simple cellular models. Overall, these findings provide novel insights into the still elusive role of CHIP in Tau pathophysiology and pave the way to new investigations on the biological significance of ubiquitination of insoluble Tau assemblies.
Acknowledgments
This work was supported by a grant from the Alzheimer's Association (AARG-17-529221 to MD). FP is recipient of a fellowship grant from the University of Verona. We acknowledge the Centro Piattaforme Tecnologiche of the University of Verona and the Electron Microscopy Facility of the University of Padova for access to the instrumentation. We thank Dr. G. Zanusso and Dr. M Fiorini (Department of Neurological and Vision Sciences, University of Verona) for the use of the FLUOstar Omega microplate reader.
Conflict of interests
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.