Birth and Future of Multiscale Modeling for Macromolecular Systems (Nobel Lecture)†
Corresponding Author
Prof. Michael Levitt
Stanford University School of Medicine, Department of Structural Biology, 299 West Campus Drive, Stanford, CA 94305 (USA)
Stanford University School of Medicine, Department of Structural Biology, 299 West Campus Drive, Stanford, CA 94305 (USA)Search for more papers by this authorCorresponding Author
Prof. Michael Levitt
Stanford University School of Medicine, Department of Structural Biology, 299 West Campus Drive, Stanford, CA 94305 (USA)
Stanford University School of Medicine, Department of Structural Biology, 299 West Campus Drive, Stanford, CA 94305 (USA)Search for more papers by this authorCopyright© The Nobel Foundation 2013. We thank the Nobel Foundation, Stockholm, for permission to print this lecture.
Graphical Abstract
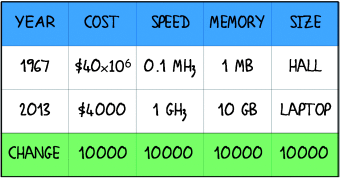
The computer industry should have received a share of the 2013 Nobel prize in chemistry as its massive research and development efforts led to unimaginable gains in computer speed (see table). This means that the cost of a particular calculation today is 100 000 000 less than it was at the beginning of Michael Levitt's scientific career as pointed out in his very personal account on the occasion of having received the Nobel prize.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie_201403691_sm_fig_14.mpg18.2 MB | fig_14 |
| anie_201403691_sm_fig_15.mpg21.6 MB | fig_15 |
| anie_201403691_sm_fig_16.mpg27.7 MB | fig_16 |
| anie_201403691_sm_fig_17a.mpg6.9 MB | fig_17a |
| anie_201403691_sm_fig_17b.mpg10.8 MB | fig_17b |
| anie_201403691_sm_miscellaneous_information.pdf58.6 KB | miscellaneous_information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1“Molecular Structure for Nucleic Acids”: J. D. Watson, F. H. C. Crick, Nature 1953, 171, 737–738.
- 2“Molecular Structure of Deoxypentose Nucleic Acids”: M. H. F. Wilkins, A. R. Stokes, H. R. Wilson, Nature 1953, 171, 738–740.
- 3“Structure of Myoglobin: A Three-Dimensional Fourier Synthesis at 2 Å Resolution”: J. C. Kendrew, R. E. Dickerson, B. E. Strandberg, R. G. Hart, D. R. Davies, D. C. Phillips, V. C. Shore, Nature 1960, 185, 422–427.
- 4“The Three-Dimensional Structure of a Protein Molecule”: J. C. Kendrew, Sci. Am. 1961, 205, 96–110.
- 5“The Structure of Haemoglobin. IV. Sign Determination by the Isomorphous Replacement Method”: D. W. Green, V. M. Ingram, M. F. Perutz, Proc. R. Soc. London Ser. A 1954, 225, 287–307.
- 6“Structure of Haemoglobin: A Three-Dimensional Fourier synthesis at 5.5 Å Resolution, Obtained by X-ray Analysis”: M. F. Perutz, M. G. Rossmann, A. F. Cullis, H. Muirhead, G. Will, A. C. T. North, Nature 1960, 185, 3416–3422.
- 7“The Hen Egg-White Lysozyme Molecule”: D. C. Phillips, Proc. Natl. Acad. Sci. USA 1967, 57, 483–495.
- 8“The Three-Dimensional Strcuture of an Enzyme Molecule”: D. C. Phillips, Sci. Am. 1966, 205, 96–110.
- 9“A Consistent Force Field for Calculation on Conformations, Vibrational Spectra and Enthalpies of Cycloalkanes and n-Alkane Molecules”: S. Lifson, A. Warshel, J. Phys. Chem. 1968, 49, 5116–5129.
- 10“A Quantum Mechanical Polarizable Force Field for Biomolecular Interactions”: A. G. Donchev, V. D. Ozrin, M. V. Subbotin, O. V. Tarasov, V. I. Tarasov, Proc. Natl. Acad. Sci. USA 2005, 102, 7829–7834.
- 11“Refinement of Protein Conformations Using a Macromolecular Energy Minimization Procedure”: M. Levitt, S. Lifson, J. Mol. Biol. 1969, 46, 269–279.
- 12“Correlations in the Motion of Atoms in Liquid Argon”: A. Rahman, Phys. Rev. 1964, 136, A 405–A411.
- 13“Molecular Dynamics Study of Liquid Water”: A. Rahman, F. H. Stillinger, J. Chem. Phys. 1971, 55, 3336–3359.
- 14“Computer Simulation of Protein Folding”: M. Levitt, A. Warshel, Nature 1975, 253, 694–698.
- 15“A Simplified Representation of Protein Conformations for Rapid Simulation of Protein Folding”: M. Levitt, J. Mol. Biol. 1976, 104, 59–107.
- 16“Minimization of Polypeptide Energy. I. Preliminary Structures of Bovine Pancreatic Ribonuclease S-peptide”: K. D. Gibson, H. A. Scheraga, Proc. Natl. Acad. Sci. USA 1967, 58, 420–427.
- 17“Conformation Analysis of Proteins”: M. Levitt, Ph.D. Thesis, Cambridge University, Cambridge, UK, 1971. http://csb.stanford.edu/levitt/Levitt_Thesis_1971/Levitt_Thesis_1971.html.
- 18“On the Nature of the Binding of Hexa-N-Acetyl Glucosamine Substrate to Lysozyme”: M. Levitt, Peptides, Polypeptides and Proteins, Wiley, New York, 1974, pp. 99–113.
- 19“Theoretical Studies of Enzymic Reactions: Dielectric, Electrostatic and Steric Stabilization of the Carbonium Ion in the Reaction of Lysozyme”: A. Warshel, M. Levitt, J. Mol. Biol. 1976, 103, 227–249.
- 20“Dynamics of Folded Proteins”: J. A. McCammon, B. R. Gelin, M. Karplus, Nature 1977, 267, 585–590.
- 21“Accurate Simulation of Protein Dynamics in Solution”: M. Levitt, R. Sharon, Proc. Natl. Acad. Sci. USA 1988, 85, 7557–7561.
- 22“Molecular Dynamics Simulation of Helix Denaturation”: V. Daggett, M. Levitt, J. Mol. Biol. 1992, 223, 1121–1138.
- 23“Atomistic Protein Folding Simulations on the Submillisecond Time Scale Using Worldwide Distributed Computing”: V. S. Pande, I. Baker, J. Chapman, S. P. Elmer, S. Khaliq, S. M. Larson, M. R. Shirts, C. D. Snow, E. J. Sorin, B. Zagrovic, Biopolymers 2003, 68, 91–109.
- 24“Anton: A Special-Purpose Machine for Molecular Dynamics Simulation”: D. E. Shaw, M. M. Deneroff, R. O. Dror, J. S. Kuskin, R. H. Larson, J. K. Salmon, C. Young, B. Batson, K. J. Bowers, J. C. Chao, M. P. Eastwood, J. Gagliardo, J. P. Grossman, C. R. Ho, D. J. Ierardi, I. Kolossváry, J. L. Klepeis, T. Layman, C. McLeavey, M. A. Moraes, R. Mueller, E. C. Priest, Y. Shan, J. Spengler, M. Theobald, B. Towles, S. C. Wang, Proceedings of the 34rd Annual International Symposium on Computer Architecture (ISCA), San Diego, June 2007.
- 25“The Molecular Basis of Eukaryotic Transcription (Nobel Lecture)”: R. Kornberg, Angew. Chem. 2007, 119, 7082–7092;
10.1002/ange.200701832 Google ScholarAngew. Chem. Int. Ed. 2007, 48, 6956–6965.
- 26“Automatic Discovery of Metastable States for the Construction of Markov Models Of Macromolecular Conformational Dynamics”: J. D. Chodera, N. Singhal, V. S. Pande, K. A. Dill, W. C. Swope, J. Chem. Phys. 2007, 126, 155101–17.
- 27“Can Morphing Methods Predict Intermediate Structures?”: D. Weiss, M. Levitt, J. Mol. Biol. 2009, 385, 665–674.
- 28“Millisecond Dynamics of RNA Polymerase II Translocation at Atomic Resolution”: D.-A. Silva, D. Weiss, F. Pardo, L.-T. Da, M. Levitt, D. Wang, X. Huang, Proc. Natl. Acad. Sci. USA 2014, 111, 6293–6298.
- 29“Protein Normal-Mode Dynamics: Trypsin Inhibitor, Crambin, Ribonuclease and Lysozyme”: M. Levitt, C. Sander, P. S. Stern, J. Mol. Biol. 1985, 181, 423–447.
- 30“Unraveling the Structure of the Ribosome (Nobel Lecture)”: V. Ramakrishnan, Angew. Chem. 2010, 122, 4454–4481;
10.1002/ange.201001436 Google ScholarAngew. Chem. Int. Ed. 2010, 49, 4355–4380.
- 31“From the Structure and Function of the Ribosome to New Antibiotics (Nobel Lecture)”: T. A. Steitz, Angew. Chem. 2010, 122, 4482–4500;
10.1002/ange.201000708 Google ScholarAngew. Chem. Int. Ed. 2010, 49, 4381–4398.
- 32“Hibernating Bears, Antibiotics, and the Evolving Ribosome (Nobel Lecture)”: A. Yonath, Angew. Chem. 2010, 122, 4438–4453;
10.1002/ange.201001297 Google ScholarAngew. Chem. Int. Ed. 2010, 49, 4340–4354.
- 33“Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis”: M. Tirion, Phys. Rev. Lett. 1996, 77, 1905–1908.
- 34“Conformational Optimization with Natural Degrees of Freedom: A Novel Stochastic Chain Closure Algorithm”: P. Minary, M. Levitt, J. Comput. Chem. 2010, 17, 993–1010.
- 35“Molecular Flexibility in ab initio Drug Docking to DNA: Binding-Site and Binding-Mode Transitions in All-Atom Monte Carlo Simulations”: R. Rohs, I. Bloch, H. Sklenar, Z. Shakked, Nucleic Acids Res. 2005, 33, 7048–7057.
- 36“Modeling and Design by Hierarchical Natural Moves”: A. Y. L. Sim, M. Levitt, P. Minary, Proc. Natl. Acad. Sci. USA 2012, 109, 2890–2895.
- 37“Training-free atomistic prediction of nucleosome occupancy”: P. Minary, M. Levitt, Proc. Natl. Acad. Sci. USA 2014, 111, 7665—7670.
- 38“Subunit Order of Eukaryotic TRiC/CCT Chaperonin by Cross-linking, Mass Spectrometry and Combinatorial Homology Modeling”: N. Kalisman, C. M. Adams, M. Levitt, Proc. Natl. Acad. Sci. USA 2012, 109, 2884–2889.
- 39“Architecture of an RNA polymerase II transcription pre-initiation complex”: K. Murakami, H. Elmlund, N. Kalisman, D. A. Bushnell, C. M. Adams, M. Azubel, D. Elmlund, Y. Levi-Kalisman, X. Liu, B. J. Gibbons, M. Levitt, R. D. Kornberg, Science 2013, 342, 1238724.
- 40“Application of a polarizable force field to calculations of relative protein–ligand binding affinities”: O. Khoruzhii, A. G. Donchev, G. Nikolay, A. Illarionov, M. Olevanov, V. Ozrin, C. Queen, V. Tarasov, Proc. Natl. Acad. Sci. USA 2008, 105, 10378–10383.
- 41“The Predicted Structure of Immunoglobulin D1.3 and its Comparison with the Crystal Structure”: C. Chothia, A. M. Lesk, M. Levitt, A. G. Amit, R. A. Mariuzza, S. E. V. Phillips, R. J. Poljak, Science 1986, 233, 755–758.
- 42“The Conformations of Immunoglobulin Hypervariable Regions”: C. Chothia, A. M. Lesk, M. Levitt, A. Tramontano, S. J. Smith-Gill, G. Air, S. Sheriff, E. A. Padlan, D. Davies, W. R. Tulip, P. M. Colman, Nature 1989, 342, 877–883.
- 43“A Humanized Antibody that Binds to the IL-2 Receptor”: C. Queen, W. P. Schneider, H. E. Selick, P. W. Payne, N. F. Landolfi, J. F. Duncan, A. M. Avdalovic, M. Levitt, R. P. Junghans, T. A. Waldmann, Proc. Natl. Acad. Sci. USA 1989, 86, 10029–10033.
- 44“Replacing the Complementarity-Determining Regions in a Human Antibody with those from a Mouse”: P. T. Jones, P. H. Dear, J. Foote, M. S. Neuberger, G. Winter, Nature 1986, 321, 522–525.
- 45“Michael Levitt National Academy of Science USA Member Details”: http://nrc88.nas.edu/pnas_search/memberDetails.aspx?ctID=3012570.
- 46“Life-Long Influence of Max Perutz and the Laboratory of Molecular Biology”: M. Levitt in Memories and Consequences (Ed.: H. Huxley), 2013. http://www2.mrc-lmb.cam.ac.uk/ebooks/Memories_and_consequences-Hugh_Huxley.epub.