Single-Molecule Analysis of the Recognition Forces Underlying Nucleo-Cytoplasmic Transport†
Dr. Martina Rangl
Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorProf. Andreas Ebner
Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorJustin Yamada
Department of Molecular, Cell and Developmental Biology, Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064 (USA)
Search for more papers by this authorDr. Christian Rankl
Agilent Technologies Austria GmbH, Mooslackengasse 17, 1190 Wien (Austria)
Search for more papers by this authorProf. Robert Tampé
Institute of Biochemistry, Biocenter, Goethe-University Frankfurt, Frankfurt (Germany)
Search for more papers by this authorProf. Hermann J. Gruber
Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorCorresponding Author
Prof. Michael Rexach
Department of Molecular, Cell and Developmental Biology, Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064 (USA)
Michael Rexach, Department of Molecular, Cell and Developmental Biology, Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064 (USA)
Peter Hinterdorfer, Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorCorresponding Author
Prof. Peter Hinterdorfer
Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Michael Rexach, Department of Molecular, Cell and Developmental Biology, Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064 (USA)
Peter Hinterdorfer, Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorDr. Martina Rangl
Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorProf. Andreas Ebner
Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorJustin Yamada
Department of Molecular, Cell and Developmental Biology, Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064 (USA)
Search for more papers by this authorDr. Christian Rankl
Agilent Technologies Austria GmbH, Mooslackengasse 17, 1190 Wien (Austria)
Search for more papers by this authorProf. Robert Tampé
Institute of Biochemistry, Biocenter, Goethe-University Frankfurt, Frankfurt (Germany)
Search for more papers by this authorProf. Hermann J. Gruber
Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorCorresponding Author
Prof. Michael Rexach
Department of Molecular, Cell and Developmental Biology, Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064 (USA)
Michael Rexach, Department of Molecular, Cell and Developmental Biology, Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064 (USA)
Peter Hinterdorfer, Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorCorresponding Author
Prof. Peter Hinterdorfer
Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Michael Rexach, Department of Molecular, Cell and Developmental Biology, Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064 (USA)
Peter Hinterdorfer, Institute for Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz (Austria)
Search for more papers by this authorThis work was supported by NIH (RO1 grant number GM077520 to M.R.), the Austrian Science Fund (grant number FWF W1201 N13 to P.H.), L’Oréal Austria Scholarship “For Women in Science” to P.H., Johannes Kepler University Scholarship for young scientists to P.H., and Austrian FFG-MNT-ERA.NET Project IntelliTip (grant number 823980 to A.E.).
Graphical Abstract
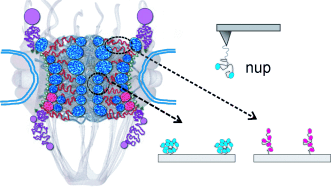
Transport forces: To move molecules across the nuclear envelope they have to overcome the selective barrier of the nuclear pore which is formed by nucleoporins (nups) with FG repeats. The molecules are chaperoned by shuttling receptors that interact with FG nups thereby passing the barrier using an unresolved mechanism. This process is explored by single-molecule force spectroscopy (see picture).
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie_201305359_sm_miscellaneous_information.pdf1.6 MB | miscellaneous_information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1C. M. Feldherr, E. Kallenbach, N. Schultz, J. Cell Biol. 1984, 99, 2216–2222.
- 2A. Hoelz, E. Debler, G. Blobel, Annu. Rev. Biochem. 2011, 80, 613–643.
- 3M. P. Rout, J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, B. T. Chait, J. Cell Biol. 2000, 148, 635–651.
- 4J. M. Cronshaw, A. N. Krutchinsky, W. Zhang, B. T. Chait, M. J. Matunis, J. Cell Biol. 2002, 158, 915–927.
- 5S. S. Patel, B. Belmont, J. Sante, M. Rexach, Cell 2007, 129, 83–96.
- 6B. Naim, D. Zbaida, S. Dagan, R. Kapon, Z. Reich, EMBO J. 2009, 28, 2697–2705.
- 7K. Ribbeck, D. Gorlich, EMBO J. 2002, 21, 2664–2671.
- 8D. Görlich, U. Kutay, Annu. Rev. Cell Dev. Biol. 1999, 15, 607–660.
- 9Y. Chook, K. Suël, Biochim. Biophys. Acta 2010, E pub ahead of print.
- 10S. Wente, M. Rout, Cold Spring Harbor Perspect. Biol. 2010, 2, p. a000562. Epub.
- 11D. P. Denning, S. S. Patel, V. Uversky, A. L. Fink, M. Rexach, Proc. Natl. Acad. Sci. USA 2003, 100, 2450–2455.
- 12F. Alber et al., Nature 2007, 450, 695–701.
- 13J. Yamada et al., Mol. Cell. Proteomics 2010, 9, 2205–2224.
- 14A. Radu, M. Moore, G. Blobel, Cell 1995, 81, 215–222.
- 15R. Bayliss, T. Littlewood, L. A. Strawn, S. R. Wente, M. Stewart, J. Biol. Chem. 2002, 277, 50597–50606.
- 16D. Denning, M. Rexach, Mol. Cell. Proteomics 2007, 6, 272–282.
- 17K. Ribbeck, D. Gorlich, EMBO J. 2001, 20, 1320–1330.
- 18R. Peters, Traffic 2005, 6, 421–427.
- 19E. L. Florin, V. T. Moy, H. E. Gaub, Science 1994, 264, 415–417.
- 20P. Hinterdorfer, W. Baumgartner, H. J. Gruber, K. Schilcher, H. Schindler, Proc. Natl. Acad. Sci. USA 1996, 93, 3477.
- 21S. Otsuka, S. Iwasaka, Y. Yoneda, K. Takeyasu, S. Yoshimura, Proc. Natl. Acad. Sci. USA 2008, 105, 16101–16106.
- 22R. Nevo, C. Stroh, F. Kienberger, D. Kaftan, V. Brumfeld, M. Elbaum, Z. Reich, P. Hinterdorfer, Nat. Struct. Biol. 2003, 10, 553–557.
- 23R. Nevo, V. Brumfeld, M. Elbaum, P. Hinterdorfer, Z. Reich, Biophys. J. 2004, 87, 4, 2630–2634.
- 24R. Nevo, V. Brumfeld, R. Kapon, P. Hinterdorfer, Z. Reich, EMBO Rep. 2005, 6, 482–486.
- 25R. Y. Lim, B. Fahrenkrog, J. Köser, K. Schwarz-Herion, J. Deng, U. Aebi, Science 2007, 318, 640–643.
- 26V. Krishnan, E. Lau, J. Yamada, D. Denning, S. Patel, M. Colvin, M. Rexach, PLoS Comput. Biol. 2008, 4, e 1000145.
- 27T. A. Isgro, K. Schulten, Structure 2005, 13, 1869–1879.
- 28C. Müller, G. Cingolani, C. Petosa, K. Weis, Nature 1999, 399, 221–229.
- 29E. Evans, K. Ritchie, Biophys. J. 1997, 72, 1541–1555.
- 30C. Rankl, F. Kienberger, L. Wildling, J. Wruss, H. J. Gruber, D. Blaas, P. Hinterdorfer, Proc. Natl. Acad. Sci. USA 2008, 105, 17778–17783.
- 31W. Yang, S. Musser, J. Cell Biol. 2006, 174, 951–961.
- 32R. L. Schoch, L. E. Kapinos, R. Y. Lim, Proc. Natl. Acad. Sci. USA 2012, 109, 16911–16916.
- 33C. Ader, S. Frey, W. Maas, H. Schmidt, D. Gorlich, M. Baldus, Proc. Natl. Acad. Sci. USA 2010, 107, 6281–6285.
- 34S. Alberti, R. Halfmann, O. King, A. Kapila, S. Lindquist, Cell 2009, 137, 146–158.