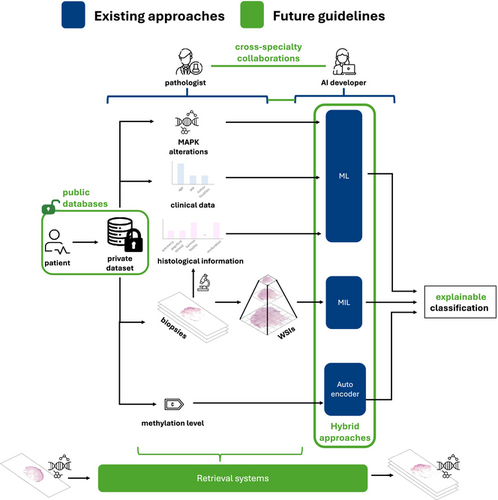
The puzzling Spitz tumours: is artificial intelligence the key to their understanding?
Corresponding Author
Laëtitia Launet
Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain
Address for correspondence: Laëtitia Launet and Valery Naranjo, Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain. e-mail: [email protected] and [email protected]
Search for more papers by this authorAdrián Colomer
Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain
Search for more papers by this authorAndrés Mosquera-Zamudio
Universitat de València, Valencia, Spain
INCLIVA, Instituto de Investigación Sanitaria, Valencia, Spain
Search for more papers by this authorCarlos Monteagudo
Universitat de València, Valencia, Spain
INCLIVA, Instituto de Investigación Sanitaria, Valencia, Spain
Search for more papers by this authorCorresponding Author
Valery Naranjo
Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain
Address for correspondence: Laëtitia Launet and Valery Naranjo, Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain. e-mail: [email protected] and [email protected]
Search for more papers by this authorCorresponding Author
Laëtitia Launet
Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain
Address for correspondence: Laëtitia Launet and Valery Naranjo, Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain. e-mail: [email protected] and [email protected]
Search for more papers by this authorAdrián Colomer
Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain
Search for more papers by this authorAndrés Mosquera-Zamudio
Universitat de València, Valencia, Spain
INCLIVA, Instituto de Investigación Sanitaria, Valencia, Spain
Search for more papers by this authorCarlos Monteagudo
Universitat de València, Valencia, Spain
INCLIVA, Instituto de Investigación Sanitaria, Valencia, Spain
Search for more papers by this authorCorresponding Author
Valery Naranjo
Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain
Address for correspondence: Laëtitia Launet and Valery Naranjo, Instituto Universitario de Investigación en Tecnología Centrada en el Ser Humano, HUMAN-Tech, Universitat Politècnica de València, Valencia, Spain. e-mail: [email protected] and [email protected]
Search for more papers by this authorAbstract
Since their first description in 1948, Spitz tumours remain one of the most challenging diagnostic entities in dermatopathology due to their complex histological features and ambiguous clinical behaviour. In recent years, artificial intelligence (AI) solutions have demonstrated significant potential across a wide range of medical applications, including computational pathology, for decision-making in diagnosis, along with promising advances in prognosis and tumour classification. However, the application of AI to Spitz tumours remains relatively underexplored, with few studies addressing this field. Yet in this evolving technological landscape, could AI provide the insights needed to help resolve the diagnostic uncertainties surrounding Spitz tumours? How could this technology be leveraged to bridge the gap between histopathological uncertainty and clinical accuracy? This review aims to provide an overview of the current state of AI applications in Spitz tumour analysis, identify existing research gaps, and propose future directions to optimize the use of AI in understanding and diagnosing these complex tumours.
Graphical Abstract
Conflict of interest
The authors declare no conflicts of interest. The founders had no role in the study's design; in the collection, analysis, or interpretation of data; in the writing of the article, or in the decision to publish the results.
Open Research
Data availability statement
Data sharing was not applicable to this article as no datasets were generated or analysed during the current study.
References
- 1Spitz S. Melanomas of childhood. Am. J. Pathol. 1948; 24; 591.
- 2Lallas A, Kyrgidis A, Ferrara G et al. Atypical Spitz tumours and sentinel lymph node biopsy: a systematic review. Lancet Oncol. 2014; 15; e178–e183.
- 3Murali R, Sharma RN, Thompson JF et al. Sentinel lymph node biopsy in histologically ambiguous melanocytic tumors with spitzoid features (so-called atypical spitzoid tumors). Ann. Surg. Oncol. 2008; 15; 302–309.
- 4Massi D, De Giorgi V, Mandalà M. The complex management of atypical Spitz tumours. Pathology 2016; 48; 132–141.
- 5Yeh I, Busam KJ. Spitz melanocytic tumours–a review. Histopathology 2022; 80; 122–134.
- 6Smith KJ, Barrett TL, Skelton HG III, Lupton GP, Graham JH. Spindle cell and epithelioid cell nevi with atypia and metastasis (malignant Spitz nevus). Am. J. Surg. Pathol. 1989; 13; 931–939.
- 7Barnhill RL, Flotte TJ, Fleischli M, Perez-Atayde A. Cutaneous melanoma and atypical Spitz tumors in childhood. Cancer 1995; 76; 1833–1845.
10.1002/1097-0142(19951115)76:10<1833::AID-CNCR2820761024>3.0.CO;2-L CAS PubMed Web of Science® Google Scholar
- 8Wiesner T, Kutzner H, Cerroni L, Mihm MC Jr, Busam KJ, Murali R. Genomic aberrations in spitzoid melanocytic tumours and their implications for diagnosis, prognosis and therapy. Pathology 2016; 48; 113–131.
- 9Rodriguez JPM, Rodriguez R, Silva VWK et al. Artificial intelligence as a tool for diagnosis in digital pathology whole slide images: a systematic review. J. Pathol. Inform. 2022; 13; 100138.
- 10Kanwal N, López-Pérez M, Kiraz U, Zuiverloon TC, Molina R, Engan K. Are you sure it's an artifact? Artifact detection and uncertainty quantification in histological images. Comput. Med. Imaging Graph. 2024; 112; 102321.
- 11Ko YS, Choi YM, Kim M et al. Improving quality control in the routine practice for histopathological interpretation of gastrointestinal endoscopic biopsies using artificial intelligence. PLoS One 2022; 17; e0278542.
- 12Mosquera-Zamudio A, Launet L, Colomer A et al. Histological interpretation of spitzoid tumours: an extensive machine learning-based concordance analysis for improving decision making. Histopathology 2024; 85(1); 155–170.
- 13JCC X-J, Ocanha-Xavier JP, WHO. 2018 classification of skin tumors. Philadelphia, United States. Am J Clin Dermatol 2019; 41; 699–700.
- 14Gerami P, Bahrami A, Busam K, de la Fouchardière A, Kazakov D, Dea M. Spitz melanoma. In D Elder, R Barnhill eds. WHO classification of tumours. Vol. 12. 5th ed. Lyon, France: International Agency for Research on Cancer, 2023. https://tumourclassification.iarc.who.int/chapters/64.
- 15Hagstrom M, Fumero-Velázquez M, Dhillon S, Olivares S, Gerami P. An update on genomic aberrations in Spitz naevi and tumours. Pathology 2023; 55; 196–205.
- 16Gerami P, Chen A, Sharma N et al. BRAF mutated and morphologically Spitzoid tumors, a subgroup of melanocytic neoplasms difficult to distinguish from true Spitz neoplasms. Am. J. Surg. Pathol. 2024; 48; 538–545.
- 17Barnhill R, Bahrami A, Bastian B et al. Malignant Spitz Tumour (Spitz Melanoma). Lyon, France: World health Organization, 2018; 108–110.
- 18Tayebi RM, Mu Y, Dehkharghanian T et al. Automated bone marrow cytology using deep learning to generate a histogram of cell types. Commun. Med. 2022; 2; 45.
- 19Feng R, Liu X, Chen J, Chen DZ, Gao H, Wu J. A deep learning approach for colonoscopy pathology WSI analysis: accurate segmentation and classification. IEEE J. Biomed. Health Inform. 2020; 25; 3700–3708.
- 20Han C, Lin J, Mai J et al. Multi-layer pseudo-supervision for histopathology tissue semantic segmentation using patch-level classification labels. Med. Image Anal. 2022; 80; 102487.
- 21Srinidhi CL, Ciga O, Martel AL. Deep neural network models for computational histopathology: a survey. Med. Image Anal. 2021; 67; 101813.
- 22Ilse M, Tomczak J, Welling M. Attention-based deep multiple instance learning. In Proceedings of the 35th International Conference on Machine Learning Proceedings of Machine Learning Research (PMLR). Stockholm, Sweden: International Conference on Machine Learning PMLR, 2018; 2127–2136.
- 23Lu MY, Williamson DF, Chen TY, Chen RJ, Barbieri M, Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021; 5; 555–570.
- 24Fernandez-Martín C, Silva-Rodriguez J, Kiraz U, Morales S, Janssen EA, Naranjo V. Uninformed teacher-student for hard-samples distillation in weakly supervised mitosis localization. Comput. Med. Imaging Graph. 2024; 112; 102328.
- 25Boyd J, Villa I, Mathieu MC et al. Region-guided cyclegans for stain transfer in whole slide images. In International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham, Switzerland: Springer Nature Switzerland. 2022; 356–365.
- 26Li J, Li W, Sisk A et al. A multi-resolution model for histopathology image classification and localization with multiple instance learning. Comput. Biol. Med. 2021; 131; 104253.
- 27Chen RJ, Lu MY, Williamson DF et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell 2022; 40; 865–878.
- 28Carvalho ED, Antonio Filho O, Silva RR et al. Breast cancer diagnosis from histopatho-logical images using textural features and CBIR. Artif. Intell. Med. 2020; 105; 101845.
- 29Wang Y, Acs B, Robertson S et al. Improved breast cancer histological grading using deep learning. Ann. Oncol. 2022; 33; 89–98.
- 30Mosquera-Zamudio A, Launet L, Tabatabaei Z et al. Deep learning for skin melanocytic tumors in whole-slide images: a systematic review. Cancer 2022; 15; 42.
10.3390/cancers15010042 Google Scholar
- 31Belaala A, Terrissa LS, Zerhouni N, Devalland C. Computer-aided diagnosis for spitzoid lesions classification using artificial intelligence techniques. Int. J. Healthc. Inf. Syst. Inform. 2021; 16; 16–37.
- 32Del Amor R, Launet L, Colomer A et al. An attention-based weakly supervised framework for spitzoid melanocytic lesion diagnosis in whole slide images. Artif. Intell. Med. 2021; 121; 102197.
- 33Launet L, Colomer A, Mosquera-Zamudio A, Moscardó A, Monteagudo C, Naranjo V. A self-training weakly-supervised framework for pathologist-like histopathological image analysis. In 2022 IEEE International Conference on Image Processing (ICIP) IEEE. Piscataway, NJ, USA: IEEE, 2022; 3401–3405.
- 34Del Amor R, Curieses FJ, Launet L et al. Multi-resolution framework for spitzoid neoplasm classification using histological data. In 2022 IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP), IEEE. Piscataway, NJ, USA: IEEE, 2022; 1–5.
- 35Tabatabaei Z, Colomer A, Moll JO, Naranjo V. Siamese content-based search engine for a more transparent skin and breast cancer diagnosis through histological imaging. arXiv preprint arXiv: 2401082722024.
- 36Del Amor R, Colomer A, Monteagudo C, Garzón MJ, García-Giménez JL, Naranjo V. A deep embedded framework for Spitzoid neoplasm classification using DNA methylation data. In 2021 29th European signal processing conference (EUSIPCO) IEEE, 2021; 1271–1275.
10.23919/EUSIPCO54536.2021.9616137 Google Scholar
- 37Lang UE, Torres R, Cheung C et al. Ciliation index is a useful diagnostic tool in challenging spitzoid melanocytic neoplasms. J. Invest. Dermatol. 2020; 140; 1401–1409.
- 38Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv: 14091556 2014.
- 39Hu J, Shen L, Sun G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Piscataway, NJ, USA: IEEE, 2018; 7132–7141.
10.1109/CVPR.2018.00745 Google Scholar
- 40Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In Medical image computing and computer-assisted intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18 Springer. Cham, Switzerland: Springer Nature Switzerland, 2015; 234–241.
- 41He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Piscataway, NJ, USA: IEEE, 2016; 770–778.
10.1109/CVPR.2016.90 Google Scholar
- 42Kim J, Dabiri S, Seeley ES. Primary cilium depletion typifies cutaneous melanoma in situ and malignant melanoma. PLoS One 2011; 6; e27410.
- 43Elmore JG, Barnhill RL, Elder DE et al. Pathologists' diagnosis of invasive melanoma and melanocytic proliferations: observer accuracy and reproducibility study. BMJ 2017; 357; j2813.
- 44Bulten W, Kartasalo K, Chen PHC et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat. Med. 2022; 28; 154–163.
- 45Spanhol FA, Oliveira LS, Petitjean C, Heutte L. A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 2015; 63; 1455–1462.
- 46Mosquera-Zamudio A, Launet L, Del Amor R et al. A Spitzoid tumor dataset with clinical metadata and whole slide images for deep learning models. Scientific Data 2023; 10; 704.
- 47Lu MY, Chen RJ, Kong D et al. Federated learning for computational pathology on gigapixel whole slide images. Med. Image Anal. 2022; 76; 102298.
- 48Launet L, Wang Y, Colomer A et al. Federating medical deep learning models from private Jupyter notebooks to distributed institutions. Appl. Sci. 2023; 13; 919.
- 49Dippel J, Feulner B, Winterhoff T et al. RudolfV: a foundation model by pathologists for pathologists. arXiv preprint arXiv:2401040792024.
- 50Huang Z, Bianchi F, Yuksekgonul M, Montine TJ, Zou J. A visual–language foundation model for pathology image analysis using medical twitter. Nat. Med. 2023; 29; 2307–2316.
- 51Lu MY, Chen B, Williamson DF et al. A visual-language foundation model for computational pathology. Nat. Med. 2024; 30; 863–874.
- 52March J, Hand M, Truong A, Grossman D. Practical application of new technologies for melanoma diagnosis: Part II. Molecular approaches. J. Am. Acad. Dermatol. 2015; 72; 943–958.
- 53Bonilla JM, Tabanera MT, Mendoza LR. El cáncer de mama en el siglo XXI: de la detección precoz a los nuevos tratamientos. Radiologia 2017; 59; 368–379.
- 54Pulgarín-Ospina CC, del Amor R, Colomera A, Silva-Rodríguez J, Naranjo V. Histocolai: an open-source web platform for collaborative digital histology image annotation with ai-driven predictive integration. arXiv preprint arXiv:2307075252023.
- 55Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In I Guyon, UV Luxburg, S Bengio et al. eds. Advances in Neural Information Processing Systems. Vol. 30. Red Hook, NY, USA: Curran Associates, Inc, 2017. https://proceedings.neurips.cc/paper_files/paper/2017/file/8a20a8621978632d76c43dfd28b67767-Paper.pdf.
- 56Del Amor R, Pérez-Cano J, López-Pérez M et al. Annotation protocol and crowdsourcing multiple instance learning classification of skin histological images: the CR-AI4SkIN dataset. Artif. Intell. Med. 2023; 145; 102686.
Online Version of Record before inclusion in an issue