Impact of hydroxycarbamide treatment on the whole-blood transcriptome in sickle cell disease
Summary
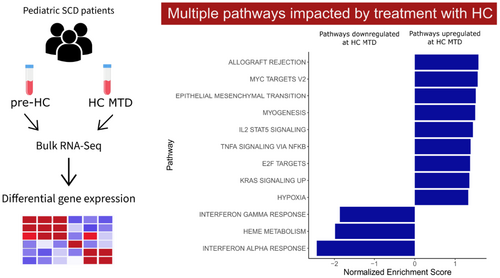
Hydroxycarbamide (HC) is the most widely used therapeutic for individuals with sickle cell disease (SCD, including sickle cell anemia and other forms of the disease). HC's clinical benefits are primarily associated with its ability to induce foetal haemoglobin (HbF); this limited view of HC's therapeutic potential may lead to its discontinuation when a modest amount of HbF is induced. A better understanding of the HbF-independent effects of HC on genes and pathways relevant to SCD pathophysiology is therefore needed. In this study, we performed bulk RNA-Seq on whole blood samples collected from a cohort of 25 paediatric patients with SCD to identify genes and pathways that are affected by treatment with HC. At the maximum tolerated dose (MTD) of HC, patients showed altered expression levels of several genes and biological pathways. Pathways related to haeme metabolism, interferon-alpha response, and interferon-gamma response were significantly downregulated at HC MTD relative to the matched pre-HC samples. Pathways linked with IL2-STAT5 signalling and TNFα signalling via NF-Kβ were observed to be up-regulated at HC MTD. These results illustrate the range of effects exerted by HC during therapy for SCD and pave the way for an improved understanding of the HbF induction-independent benefits of HC.
Graphical Abstract
Hydroxycarbamide (HC) is the most widely used therapeutic for individuals with sickle cell disease (SCD). HC's benefits are primarily associated with its ability to induce foetal haemoglobin (HbF); this limited view of HC's therapeutic potential may lead to its discontinuation when a modest amount of HbF is induced. Here, we performed bulk RNA-Seq on whole blood samples collected from 25 paediatric patients with SCD to identify genes and pathways affected by treatment with HC. Pathways related to haeme metabolism, interferon-gamma response, and interferon-alpha response were significantly downregulated at HC MTD relative to the matched pre-HC samples. Pathways linked with IL2-STAT5 signalling and TNFα signalling via NF-Kβ were observed to be up-regulated at HC MTD. These results pave the way for an improved understanding of the HbF induction-independent benefits of HC.
CONFLICT OF INTEREST STATEMENT
Vivien A. Sheehan receives research funding from Beam Therapeutics, Pfizer, Novartis, and Agios. Alka A. Potdar and G. Karen Yu are former employees of Pfizer.
Open Research
DATA AVAILABILITY STATEMENT
The bulk RNA-Seq data generated for this study is available at GEO under accession number GSE254951.