Mechanistic Study of Oxidoreductase AprQ Involved in Biosynthesis of Aminoglycoside Antibiotic Apramycin
Jinxiu Wang
State Key Laboratory of Cryospheric Science, Key Laboratory of Extreme Environmental Microbial Resources and Engineering, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu, 730000 China
Department of Chemistry, Fudan University, Shanghai, 200433 China
University of Chinese Academy of Sciences, Beijing, 100049 China
Search for more papers by this authorSuze Ma
Department of Chemistry, Fudan University, Shanghai, 200433 China
Search for more papers by this authorWei Ding
State Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240 China
Search for more papers by this authorCorresponding Author
Tuo Chen
State Key Laboratory of Cryospheric Science, Key Laboratory of Extreme Environmental Microbial Resources and Engineering, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu, 730000 China
E-mail: [email protected]; [email protected]Search for more papers by this authorCorresponding Author
Qi Zhang
Department of Chemistry, Fudan University, Shanghai, 200433 China
E-mail: [email protected]; [email protected]Search for more papers by this authorJinxiu Wang
State Key Laboratory of Cryospheric Science, Key Laboratory of Extreme Environmental Microbial Resources and Engineering, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu, 730000 China
Department of Chemistry, Fudan University, Shanghai, 200433 China
University of Chinese Academy of Sciences, Beijing, 100049 China
Search for more papers by this authorSuze Ma
Department of Chemistry, Fudan University, Shanghai, 200433 China
Search for more papers by this authorWei Ding
State Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240 China
Search for more papers by this authorCorresponding Author
Tuo Chen
State Key Laboratory of Cryospheric Science, Key Laboratory of Extreme Environmental Microbial Resources and Engineering, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu, 730000 China
E-mail: [email protected]; [email protected]Search for more papers by this authorCorresponding Author
Qi Zhang
Department of Chemistry, Fudan University, Shanghai, 200433 China
E-mail: [email protected]; [email protected]Search for more papers by this authorMain observation and conclusion
The aminoglycoside antibiotic apramycin contains a unique bicyclic octose moiety, and biosynthesis of this moiety involves an oxidoreductase AprQ. Unlike other known “Q” series proteins involved in aminoglycosides biosynthesis, AprQ does not work with an aminotransferase partner, and performs a four-electron oxidation that converts a CH2OH moiety to a carboxylate group. In this study, we report mechanistic investigation of AprQ. We showed AprQ contains a flavin mononucleotide (FMN) cofactor, which is different from other known Q series enzymes that contain a flavin adenine dinucleotide (FAD) cofactor. A series of biochemical assays showed that AprQ is not a monooxygenase but a flavoprotein oxidase. Although molecular O2 is strictly required for reaction turnover, four-electron oxidation can be achieved in the absence of O2 in single turnover condition. These findings establish the detailed catalytic mechanism of AprQ and expand the growing family of flavoprotein oxidases, an increasingly important class of biocatalysts.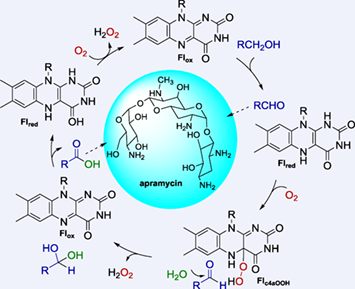
Supporting Information
| Filename | Description |
|---|---|
| cjoc202100070-sup-0001-Supinfo.pdfPDF document, 842 KB |
Appendix S1: Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1 O'Connor, S.; Lam, L. K.; Jones, N. D.; Chaney, M. O. Apramycin, a unique aminocyclitol antibiotic. J. Org. Chem. 1976, 41, 2087–2092.
- 2 Kudo, F.; Eguchi, T. Biosynthetic genes for aminoglycoside antibiotics. J. Antibiot. 2009, 62, 471–481.
- 3 Lin, C. I.; McCarty, R. M.; Liu, H. W. The biosynthesis of nitrogen-, sulfur-, and high-carbon chain-containing sugars. Chem. Soc. Rev. 2013, 42, 4377–4407.
- 4 Ni, X.; Li, D.; Yang, L.; Huang, T.; Li, H.; Xia, H. Construction of kanamycin B overproducing strain by genetic engineering of Streptomyces tenebrarius. Appl. Microbiol. Biotechnol. 2011, 89, 723–731.
- 5 Kim, H. J.; LeVieux, J.; Yeh, Y. C.; Liu, H. W. C3'-Deoxygenation of Paromamine Catalyzed by a Radical S-Adenosylmethionine Enzyme: Characterization of the Enzyme AprD4 and Its Reductase Partner AprD3. Angew. Chem. Int. Ed. 2016, 55, 3724–3728.
- 6 Lv, M.; Ji, X.; Zhao, J.; Li, Y.; Zhang, C.; Su, L.; Ding, W.; Deng, Z.; Yu, Y.; Zhang, Q. Characterization of a C3 Deoxygenation Pathway Reveals a Key Branch Point in Aminoglycoside Biosynthesis. J. Am. Chem. Soc. 2016, 138, 6427–6435.
- 7 Kudo, F.; Tokumitsu, T.; Eguchi, T. Substrate specificity of radical S-adenosyl-l-methionine dehydratase AprD4 and its partner reductase AprD3 in the C3'-deoxygenation of aminoglycoside antibiotics. J. Antibiot. 2017, 70, 423–428.
- 8 Liu, W. Q.; Amara, P.; Mouesca, J. M.; Ji, X.; Renoux, O.; Martin, L.; Zhang, C.; Zhang, Q.; Nicolet, Y. 1,2-Diol Dehydration by the Radical SAM Enzyme AprD4: A Matter of Proton Circulation and Substrate Flexibility. J. Am. Chem. Soc. 2018, 140, 1365–1371.
- 9 Park, S. R.; Park, J. W.; Ban, Y. H.; Sohng, J. K.; Yoon, Y. J. 2-Deoxystreptamine-containing aminoglycoside antibiotics: recent advances in the characterization and manipulation of their biosynthetic pathways. Nat. Prod. Rep. 2013, 30, 11–20.
- 10 Yu, Y.; Zhang, Q.; Deng, Z. Parallel pathways in the biosynthesis of aminoglycoside antibiotics. F1000Research 2017, 6, 723.
- 11 Park, J. W.; Ban, Y. H.; Nam, S. J.; Cha, S. S.; Yoon, Y. J. Biosynthetic pathways of aminoglycosides and their engineering. Curr. Opin. Biotechnol. 2017, 48, 33–41.
- 12 Guo, J.; Huang, F.; Huang, C.; Duan, X.; Jian, X.; Leeper, F.; Deng, Z.; Leadlay, P. F.; Sun, Y. Specificity and promiscuity at the branch point in gentamicin biosynthesis. Chem. Biol. 2014, 21, 608–618.
- 13 Gu, Y.; Ni, X.; Ren, J.; Gao, H.; Wang, D.; Xia, H. Biosynthesis of Epimers C2 and C2a in the Gentamicin C Complex. ChemBioChem 2015, 16, 1933–1942.
- 14 Eguchi, T.; Kudo, F.; Kitayama, Y.; Miyanaga, A.; Numakura, M. Stepwise Post-glycosylation Modification of Sugar Moieties in Kanamycin Biosynthesis. ChemBioChem 2021, 22, 1668–1675.
- 15 Huang, F.; Spiteller, D.; Koorbanally, N. A.; Li, Y.; Llewellyn, N. M.; Spencer, J. B. Elaboration of neosamine rings in the biosynthesis of neomycin and butirosin. ChemBioChem 2007, 8, 283–288.
- 16 Kudo, F.; Kawashima, T.; Yokoyama, K.; Eguchi, T. Enzymatic preparation of neomycin C from ribostamycin. J. Antibiot. 2009, 62, 643–646.
- 17 Clausnitzer, D.; Piepersberg, W.; Wehmeier, U. F. The oxidoreductases LivQ and NeoQ are responsible for the different 6'-modifications in the aminoglycosides lividomycin and neomycin. J. Appl. Microbiol. 2011, 111, 642–651.
- 18 Zhong, Y.; Ji, X.; Zhang, Q. Radical SAM-dependent adenosylation involved in bacteriohopanepolyol biosynthesis. Chin. J. Chem. 2020, 38, 39–42.
- 19 Zhao, J.; Ji, W.; Ji, X.; Zhang, Q. Biochemical Characterization of an Arginine 2,3-Aminomutase with Dual Substrate Specificity. Chin. J. Chem. 2020, 38, 959–962.
- 20 Cheng, J.; Ji, W.; Ma, S.; Ji, X.; Deng, Z.; Ding, W.; Zhang, Q. Characterization and Mechanistic Study of the Radical SAM Enzyme ArsS Involved in Arsenosugar Biosynthesis. Angew. Chem. Int. Ed. 2021, 60, 7570–7575.
- 21 Broderick, J. B.; Duffus, B. R.; Duschene, K. S.; Shepard, E. M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014, 114, 4229–4317.
- 22 Lanz, N. D.; Grove, T. L.; Gogonea, C. B.; Lee, K. H.; Krebs, C.; Booker, S. J. RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins. Methods. Enzymol. 2012, 516, 125–152.
- 23 Toplak, M.; Matthews, A.; Teufel, R. The devil is in the details: The chemical basis and mechanistic versatility of flavoprotein monooxygenases. Arch. Biochem. Biophys. 2021, 698, 108732.
- 24 Walsh, C. T.; Wencewicz, T. A. Flavoenzymes: versatile catalysts in biosynthetic pathways. Nat. Prod. Rep. 2013, 30, 175–200.
- 25 Martin, C.; Binda, C.; Fraaije, M. W.; Mattevi, A. The multipurpose family of flavoprotein oxidases. Enzymes 2020, 47, 63–86.
- 26 Gadda, G. Choline oxidases. Enzymes 2020, 47, 137–166.
- 27 Brodl, E.; Winkler, A.; Macheroux, P. Molecular Mechanisms of Bacterial Bioluminescence. Comput. Struct. Biotechnol. J. 2018, 16, 551–564.
- 28 Beaupre, B. A.; Moran, G. R. N5 Is the New C4a: Biochemical Functionalization of Reduced Flavins at the N5 Position. Front. Mol. Biosci. 2020, 7, 598912.
- 29 Liu, Y. C.; Li, Y. S.; Lyu, S. Y.; Hsu, L. J.; Chen, Y. H.; Huang, Y. T.; Chan, H. C.; Huang, C. J.; Chen, G. H.; Chou, C. C.; Tsai, M. D.; Li, T. L. Interception of teicoplanin oxidation intermediates yields new antimicrobial scaffolds. Nat. Chem. Biol. 2011, 7, 304–309.
- 30 Sriwaiyaphram, K.; Punthong, P.; Sucharitakul, J.; Wongnate, T. Structure and function relationships of sugar oxidases and their potential use in biocatalysis. Enzymes 2020, 47, 193–230.




