DNA Methylation Profiling Distinguishes Three Clusters of Breast Cancer Cell Lines
Abstract
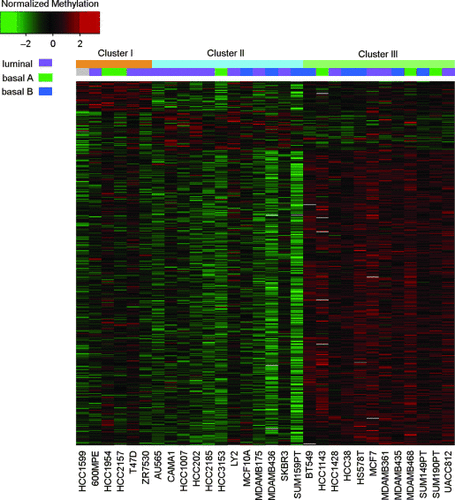
Methylation change plays an important role in many cellular systems, including cancer development. During recent years, genome-wide or large-scale methylation data has become available thanks to rapid advances in high-throughput biotechnologies. So far, researchers have always used gene expression profiling to study disease subtypes and related therapies. In this study, we investigated methylation profiles in 30 breast cancer cell lines using methylation data generated by microarray technologies. Strong variation of the number of methylation peaks was found among these 30 cell lines; however, more peaks were found in the upstream regions than in downstream regions of genes. We further grouped the methylation profiles of these cell lines into three consensus clusters. Finally, we performed an integrative analysis of breast cancer cell lines using both methylation and gene-expression profiling data. There was no significant correlation between methylation-profiling subtypes and gene-expression profiling subtypes, suggesting the complex nature of methylation in the regulation of gene expression. However, we found basal B cell lines appeared exclusively in two methylation clusters. Although these results are preliminary, this study suggests that methylation profiling might be promising in disease subtype classification and the development of therapeutic strategies.