A Computational Pipeline for Accurate Prioritization of Protein-Protein Binding Candidates in High-Throughput Protein Libraries
Dr. Arup Mondal
Department of Chemistry and Quantum Theory Project, University of Florida, Leigh Hall 240, Gainesville, FL, USA
Contribution: Conceptualization (equal), Data curation (lead), Formal analysis (lead), Investigation (lead), Methodology (lead), Validation (lead), Visualization (lead), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorBhumika Singh
Department of Chemistry and Quantum Theory Project, University of Florida, Leigh Hall 240, Gainesville, FL, USA
Contribution: Conceptualization (supporting), Data curation (supporting), Formal analysis (supporting), Investigation (supporting), Visualization (supporting)
Search for more papers by this authorRoland H. Felkner
Department of Pharmacology, Rutgers-Robert Wood Johnson Medical School, 675 Hoes Lane Rm 636, Piscataway, NJ 08854 USA
Contribution: Formal analysis (supporting), Investigation (supporting), Validation (supporting), Visualization (supporting)
Search for more papers by this authorDr. Anna De Falco
Department of Chemistry and Chemical Biology, Center for Biotechnology and Interdisciplinary Sciences, Rensselaer Polytechnic Institute, Troy, New York 12180 USA
Contribution: Formal analysis (supporting), Investigation (supporting), Validation (supporting), Visualization (supporting), Writing - original draft (supporting)
Search for more papers by this authorDr. GVT Swapna
Department of Chemistry and Chemical Biology, Center for Biotechnology and Interdisciplinary Sciences, Rensselaer Polytechnic Institute, Troy, New York 12180 USA
Contribution: Formal analysis (supporting), Investigation (supporting), Validation (supporting), Visualization (supporting)
Search for more papers by this authorCorresponding Author
Prof. Gaetano T. Montelione
Department of Chemistry and Chemical Biology, Center for Biotechnology and Interdisciplinary Sciences, Rensselaer Polytechnic Institute, Troy, New York 12180 USA
Contribution: Conceptualization (equal), Funding acquisition (equal), Project administration (equal), Resources (supporting), Supervision (equal), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorCorresponding Author
Prof. Monica J. Roth
Department of Pharmacology, Rutgers-Robert Wood Johnson Medical School, 675 Hoes Lane Rm 636, Piscataway, NJ 08854 USA
Contribution: Conceptualization (equal), Funding acquisition (equal), Investigation (equal), Project administration (equal), Resources (equal), Supervision (equal), Validation (equal), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorCorresponding Author
Prof. Alberto Perez
Department of Chemistry and Quantum Theory Project, University of Florida, Leigh Hall 240, Gainesville, FL, USA
Contribution: Conceptualization (equal), Methodology (equal), Project administration (equal), Resources (equal), Software (equal), Supervision (equal), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorDr. Arup Mondal
Department of Chemistry and Quantum Theory Project, University of Florida, Leigh Hall 240, Gainesville, FL, USA
Contribution: Conceptualization (equal), Data curation (lead), Formal analysis (lead), Investigation (lead), Methodology (lead), Validation (lead), Visualization (lead), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorBhumika Singh
Department of Chemistry and Quantum Theory Project, University of Florida, Leigh Hall 240, Gainesville, FL, USA
Contribution: Conceptualization (supporting), Data curation (supporting), Formal analysis (supporting), Investigation (supporting), Visualization (supporting)
Search for more papers by this authorRoland H. Felkner
Department of Pharmacology, Rutgers-Robert Wood Johnson Medical School, 675 Hoes Lane Rm 636, Piscataway, NJ 08854 USA
Contribution: Formal analysis (supporting), Investigation (supporting), Validation (supporting), Visualization (supporting)
Search for more papers by this authorDr. Anna De Falco
Department of Chemistry and Chemical Biology, Center for Biotechnology and Interdisciplinary Sciences, Rensselaer Polytechnic Institute, Troy, New York 12180 USA
Contribution: Formal analysis (supporting), Investigation (supporting), Validation (supporting), Visualization (supporting), Writing - original draft (supporting)
Search for more papers by this authorDr. GVT Swapna
Department of Chemistry and Chemical Biology, Center for Biotechnology and Interdisciplinary Sciences, Rensselaer Polytechnic Institute, Troy, New York 12180 USA
Contribution: Formal analysis (supporting), Investigation (supporting), Validation (supporting), Visualization (supporting)
Search for more papers by this authorCorresponding Author
Prof. Gaetano T. Montelione
Department of Chemistry and Chemical Biology, Center for Biotechnology and Interdisciplinary Sciences, Rensselaer Polytechnic Institute, Troy, New York 12180 USA
Contribution: Conceptualization (equal), Funding acquisition (equal), Project administration (equal), Resources (supporting), Supervision (equal), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorCorresponding Author
Prof. Monica J. Roth
Department of Pharmacology, Rutgers-Robert Wood Johnson Medical School, 675 Hoes Lane Rm 636, Piscataway, NJ 08854 USA
Contribution: Conceptualization (equal), Funding acquisition (equal), Investigation (equal), Project administration (equal), Resources (equal), Supervision (equal), Validation (equal), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorCorresponding Author
Prof. Alberto Perez
Department of Chemistry and Quantum Theory Project, University of Florida, Leigh Hall 240, Gainesville, FL, USA
Contribution: Conceptualization (equal), Methodology (equal), Project administration (equal), Resources (equal), Software (equal), Supervision (equal), Writing - original draft (equal), Writing - review & editing (equal)
Search for more papers by this authorGraphical Abstract
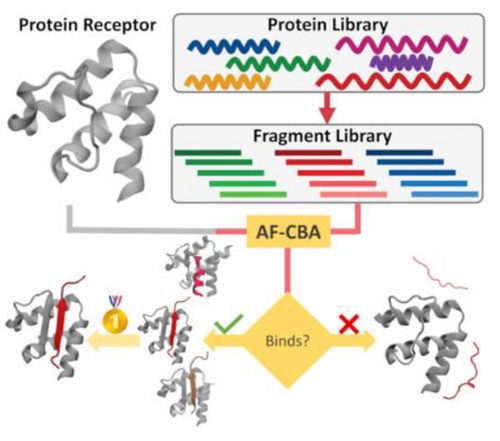
We introduce a computational pipeline that identifies peptide epitopes that bind a particular receptor from a library of possible protein sequences (e.g., coming from high-throughput experiments). The pipeline uses an AlphaFold-Competitive Binding Assay (AF-CBA) to rank order different plausible epitopes and select the most likely one.
Abstract
Identifying the interactome for a protein of interest is challenging due to the large number of possible binders. High-throughput experimental approaches narrow down possible binding partners but often include false positives. Furthermore, they provide no information about what the binding region is (e.g., the binding epitope). We introduce a novel computational pipeline based on an AlphaFold2 (AF) Competitive Binding Assay (AF-CBA) to identify proteins that bind a target of interest from a pull-down experiment and the binding epitope. Our focus is on proteins that bind the Extraterminal (ET) domain of Bromo and Extraterminal domain (BET) proteins, but we also introduce nine additional systems to show transferability to other peptide-protein systems. We describe a series of limitations to the methodology based on intrinsic deficiencies of AF and AF-CBA to help users identify scenarios where the approach will be most useful. Given the method‘s speed and accuracy, we anticipate its broad applicability to identify binding epitope regions among potential partners, setting the stage for experimental verification.
Conflict of interests
The authors declare no conflict of interest.
Open Research
Data Availability Statement
A README file, and example system, and the scripts used in this work can be downloaded from https://github.com/PDNALab/AF-CBA-pipeline. The complete results for the BRD3-MLV system can be freely downloaded from Zenodo https://doi.org/10.5281/zenodo.10801197.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie202405767-sup-0001-misc_information.pdf18.6 MB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1V. S. Rao, K. Srinivas, G. N. Sujini, G. N. S. Kumar, Int. J. Proteom. 2014, 2014, 147648.
- 2D. Younger, S. Berger, D. Baker, E. Klavins, Proc. Nat. Acad. Sci. 2017, 114, 12166–12171.
- 3A. Brückner, C. Polge, N. Lentze, D. Auerbach, U. Schlattner, Int. J. Mol. Sci. 2009, 10, 2763–2788.
- 4A. Louche, S. P. Salcedo, S. Bigot, bacterial Protein secretion systems: methods and protocols 2017, 247–255.
10.1007/978-1-4939-7033-9_20 Google Scholar
- 5T. N. Nguyen, J. A. Goodrich, Nat. Methods 2006, 3, 135–139.
- 6A. Jain, R. Liu, Y. K. Xiang, T. Ha, Nat. Protoc. 2012, 7, 445–452.
- 7R. Sanchez-Garcia, C. O. S. Sorzano, J. M. Carazo, J. Segura, Bioinformatics 2019, 35, 470–477.
- 8F. Guo, Q. Zou, G. Yang, D. Wang, J. Tang, J. Xu, BMC Bioinf. 2019, 20, 483.
- 9M. Mou, Z. Pan, Z. Zhou, L. Zheng, H. Zhang, S. Shi, F. Li, X. Sun, F. Zhu, Research 2023, 6, 0240.
- 10F. Soleymani, E. Paquet, H. Viktor, W. Michalowski, D. Spinello, Comput. Struct. Biotechnol. J. 2022, 20, 5316–5341.
- 11B. Dunham, M. K. Ganapathiraju, Molecules 2021, 27, 41.
- 12K. Krawczyk, X. Liu, T. Baker, J. Shi, C. M. Deane, Bioinformatics 2014, 30, 2288–2294.
- 13J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli, D. Hassabis, Nature 2021, DOI 10.1038/s41586-021-03819-2.
- 14L. Chang, A. Perez, Angew. Chem. Int. Ed. 2023, 62, e202213362.
- 15J. Sarnik, T. Popławski, P. Tokarz, Int. J. Mol. Sci. 2021, 22, 11102.
- 16N. Wang, R. Wu, D. Tang, R. Kang, Signal Transduct. Target. Ther. 2021, 6, 23.
- 17A. Mondal, G. V. T. Swapna, M. M. Lopez, L. Klang, J. Hao, L. Ma, M. J. Roth, G. T. Montelione, A. Perez, J. Chem. Inf. Model. 2023, 63, 2058–2072.
- 18Y. Taniguchi, Int. J. Mol. Sci. 2016, 17, 1849.
- 19S. K. Burley, C. Bhikadiya, C. Bi, S. Bittrich, H. Chao, L. Chen, P. A. Craig, G. V. Crichlow, K. Dalenberg, J. M. Duarte, S. Dutta, M. Fayazi, Z. Feng, J. W. Flatt, S. Ganesan, S. Ghosh, D. S. Goodsell, R. K. Green, V. Guranovic, J. Henry, B. P. Hudson, I. Khokhriakov, C. L. Lawson, Y. Liang, R. Lowe, E. Peisach, I. Persikova, D. W. Piehl, Y. Rose, A. Sali, J. Segura, M. Sekharan, C. Shao, B. Vallat, M. Voigt, B. Webb, J. D. Westbrook, S. Whetstone, J. Y. Young, A. Zalevsky, C. Zardecki, Nucleic Acids Res. 2022, 51, D488–D508.
- 20C. Zardecki, S. Dutta, D. S. Goodsell, M. Voigt, S. K. Burley, J. Chem. Educ. 2016, 93, 569–575.
- 21D. C. C. Wai, T. N. Szyszka, A. E. Campbell, C. Kwong, L. E. Wilkinson-White, A. P. G. Silva, J. K. K. Low, A. H. Kwan, R. Gamsjaeger, J. D. Chalmers, W. M. Patrick, B. Lu, C. R. Vakoc, G. A. Blobel, J. P. Mackay, J. Biol. Chem. 2018, 293, 7160–7175.
- 22A. L. Richards, M. Eckhardt, N. J. Krogan, Mol. Syst. Biol. 2021, 17, e8792.
- 23S. Aiyer, G. V. T. Swapna, L.-C. Ma, G. Liu, J. Hao, G. Chalmers, B. C. Jacobs, G. T. Montelione, M. J. Roth, Structure 2021, 29, 886–898.e6.
- 24Q. Zhang, L. Zeng, C. Shen, Y. Ju, T. Konuma, C. Zhao, C. R. R. Vakoc, M. M. Zhou, Structure 2016, 24, 1201–1208.
- 25T. Konuma, D. Yu, C. Zhao, Y. Ju, R. Sharma, C. Ren, Q. Zhang, M.-M. Zhou, L. Zeng, Sci. Rep. 2017, 7, 16272.
- 26W. Humphrey, A. Dalke, K. Schulten, J. Mol. Graphics 1996, 14, 33–38.
- 27A. Mondal, L. Chang, A. Perez, Qrb Discov 2022, 3, DOI 10.1017/qrd.2022.14.
- 28G. S. Ratnaparkhi, R. Varadarajan, Proteins Struct. Funct. Bioinf. 1999, 36, 282–294.
10.1002/(SICI)1097-0134(19990815)36:3<282::AID-PROT3>3.0.CO;2-F CAS PubMed Web of Science® Google Scholar
- 29B. Anil, C. Riedinger, J. A. Endicott, M. E. Noble, Acta Crystallographica Section D: Biological Crystallography 2013, 69, 1358–1366.
- 30J.-W. Wu, A. E. Cocina, J. Chai, B. A. Hay, Y. Shi, Mol. Cell 2001, 8, 95–104.
- 31I. N. Krylova, E. P. Sablin, J. Moore, R. X. Xu, G. M. Waitt, J. A. MacKay, D. Juzumiene, J. M. Bynum, K. Madauss, V. Montana, et al., Cell 2005, 120, 343–355.
- 32C. R. Grace, D. Ban, J. Min, A. Mayasundari, L. Min, K. E. Finch, L. Griffiths, N. Bharatham, D. Bashford, R. K. Guy, et al., J. Mol. Biol. 2016, 428, 1290–1303.
- 33R. Y. Samson, T. Obita, B. Hodgson, M. K. Shaw, P. L.-G. Chong, R. L. Williams, S. D. Bell, Mol. Cell 2011, 41, 186–196.
- 34E. A. Ortlund, Y. Lee, I. H. Solomon, J. M. Hager, R. Safi, Y. Choi, Z. Guan, A. Tripathy, C. R. Raetz, D. P. McDonnell, et al., Nat. Struct. Mol. Biol. 2005, 12, 357–363.
- 35D. Vucic, M. C. Franklin, H. J. Wallweber, K. Das, B. P. Eckelman, H. Shin, L. O. Elliott, S. Kadkhodayan, K. Deshayes, G. S. Salvesen, et al., Biochem. J. 2005, 385, 11–20.
- 36W. J. McGrath, J. Ding, A. Didwania, R. M. Sweet, W. F. Mangel, Biochimica et Biophysica Acta (BBA)—Proteins and Proteomics 2003, 1648, 1–11.
- 37F. Poy, M. B. Yaffe, J. Sayos, K. Saxena, M. Morra, J. Sumegi, L. C. Cantley, C. Terhorst, M. J. Eck, Mol. Cell 1999, 4, 555–561.
- 38S. A. Lambert, A. Jolma, L. F. Campitelli, P. K. Das, Y. Yin, M. Albu, X. Chen, J. Taipale, T. R. Hughes, M. T. Weirauch, Cell 2018, 172, 650–665.
- 39M. Varadi, S. Anyango, M. Deshpande, S. Nair, C. Natassia, G. Yordanova, D. Yuan, O. Stroe, G. Wood, A. Laydon, A. Žídek, T. Green, K. Tunyasuvunakool, S. Petersen, J. Jumper, E. Clancy, R. Green, A. Vora, M. Lutfi, M. Figurnov, A. Cowie, N. Hobbs, P. Kohli, G. Kleywegt, E. Birney, D. Hassabis, S. Velankar, Nucleic Acids Res. 2021, 50, D439–D444.
- 40P. D. Thomas, Methods Mol. Biol. 2016, 1446, 15–24.
10.1007/978-1-4939-3743-1_2 Google Scholar
- 41S. Rahman, M. E. Sowa, M. Ottinger, J. A. Smith, Y. Shi, J. W. Harper, P. M. Howley, Mol. Cell. Biol. 2011, 31, 2641–2652.
- 42A. Alpsoy, E. C. Dykhuizen, J. Biol. Chem. 2018, 293, 3892–3903.
- 43J.-P. Lambert, S. Picaud, T. Fujisawa, H. Hou, P. Savitsky, L. Uusküla-Reimand, G. D. Gupta, H. Abdouni, Z.-Y. Lin, M. Tucholska, J. D. R. Knight, B. Gonzalez-Badillo, N. St-Denis, J. A. Newman, M. Stucki, L. Pelletier, N. Bandeira, M. D. Wilson, P. Filippakopoulos, A.-C. Gingras, Mol. Cell 2019, 73, 621–638.e17.
- 44L. Chang, A. Mondal, A. Perez, Frontiers Bioinform 2022, 2, 1046493.
- 45M. R. Arkin, Y. Tang, J. A. Wells, Chem. Biol. 2014, 21, 1102–1114.
- 46J. L. MacCallum, A. Perez, K. A. Dill, Proc. Natl. Acad. Sci. USA 2015, 112, 6985–6990.
- 47J. A. Morrone, A. Perez, J. MacCallum, K. A. Dill, J. Chem. Theory Comput. 2017, 13, 870–876.
- 48A. Savinov, S. Swanson, A. E. Keating, G.-W. Li, bioRxiv 2023, 2023.12.19.572389.
- 49C. Y. Lee, D. Hubrich, J. K. Varga, C. Schäfer, M. Welzel, E. Schumbera, M. Djokic, J. M. Strom, J. Schönfeld, J. L. Geist, F. Polat, T. J. Gibson, C. I. K. Valsecchi, M. Kumar, O. Schueler-Furman, K. Luck, Mol. Syst. Biol. 2024, 1–23.
- 50L. Chang, A. Mondal, B. Singh, Y. Martínez-Noa, A. Perez, Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2024, 14, DOI 10.1002/wcms.1693.
- 51J. J. Perez, R. A. Perez, A. Perez, Front. Mol. Biosci. 2021, 8, 681617.
- 52J. J. Perez, Curr. Top. Med. Chem. 2018, 7, 566–590.
- 53V. Mikhaylov, C. A. Brambley, G. L. J. Keller, A. G. Arbuiso, L. I. Weiss, B. M. Baker, A. J. Levine, Structure 2024, 32, 228–241.e4.
- 54N. Hashemi, B. Hao, M. Ignatov, I. Ch Paschalidis, P. Vakili, S. Vajda, D. Kozakov, Front. Bioinform. 2023, 3, 1207380.
- 55S. A. Rettie, K. V. Campbell, A. K. Bera, A. Kang, S. Kozlov, J. D. L. Cruz, V. Adebomi, G. Zhou, F. DiMaio, S. Ovchinnikov, G. Bhardwaj, bioRxiv 2023, 2023.02.25.529956.
- 56T. A. Ramelot, J. Palmer, G. T. Montelione, G. Bhardwaj, Curr. Opin. Struct. Biol. 2023, 80, 102603.