Protecting-Group-Controlled Enzymatic Glycosylation of Oligo-N-Acetyllactosamine Derivatives
Ivan A. Gagarinov
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Search for more papers by this authorDr. Tiehai Li
Complex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, GA, 30602 USA
Search for more papers by this authorNa Wei
Complex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, GA, 30602 USA
Search for more papers by this authorJavier Sastre Toraño
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Search for more papers by this authorDr. Robert P. de Vries
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Search for more papers by this authorDr. Margreet A. Wolfert
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Complex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, GA, 30602 USA
Search for more papers by this authorCorresponding Author
Prof. Geert-Jan Boons
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Complex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, GA, 30602 USA
Department of Chemistry, University of Georgia, Athens, GA, USA
Search for more papers by this authorIvan A. Gagarinov
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Search for more papers by this authorDr. Tiehai Li
Complex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, GA, 30602 USA
Search for more papers by this authorNa Wei
Complex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, GA, 30602 USA
Search for more papers by this authorJavier Sastre Toraño
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Search for more papers by this authorDr. Robert P. de Vries
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Search for more papers by this authorDr. Margreet A. Wolfert
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Complex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, GA, 30602 USA
Search for more papers by this authorCorresponding Author
Prof. Geert-Jan Boons
Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Bijvoet Center for Biomolecular Research, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
Complex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, GA, 30602 USA
Department of Chemistry, University of Georgia, Athens, GA, USA
Search for more papers by this authorGraphical Abstract
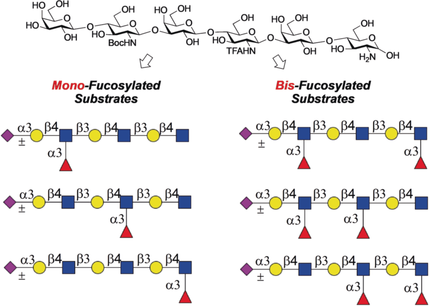
A library of differentially fucosylated and sialylated oligosaccharides was produced starting from a single chemically synthesized tri-N-acetyllactosamine derivative. The resulting oligosaccharides were printed as a microarray that was probed by several glycan-binding proteins, which showed that complex patterns of fucosylation can modulate glycan recognition.
Abstract
We describe a chemoenzymatic strategy that can give a library of differentially fucosylated and sialylated oligosaccharides starting from a single chemically synthesized tri-N-acetyllactosamine derivative. The common precursor could easily be converted into 6 different hexasaccharides in which the glucosamine moieties are either acetylated (GlcNAc) or modified as a free amine (GlcNH2) or Boc (GlcNHBoc). Fucosylation of the resulting compounds by a recombinant fucosyl transferase resulted in only modification of the natural GlcNAc moieties, providing access to 6 selectively mono- and bis-fucosylated oligosaccharides. Conversion of the GlcNH2 or GlcNHBoc moieties into the natural GlcNAc, followed by sialylation by sialyl transferases gave 12 differently fucosylated and sialylated compounds. The oligosaccharides were printed as a microarray that was probed by several glycan-binding proteins, demonstrating that complex patterns of fucosylation can modulate glycan recognition.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie201903140-sup-0001-misc_information.pdf57.3 MB | Supplementary |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1
- 1aG. W. Hart, R. J. Copeland, Cell 2010, 143, 672–676;
- 1bA. Varki, Glycobiology 2017, 27, 3–49.
- 2
- 2aT. Buskas, S. Ingale, G. J. Boons, Glycobiology 2006, 16, 113R–136R;
- 2bK. W. Moremen, M. Tiemeyer, A. V. Nairn, Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462.
- 3M. Nonaka, M. Fukuda in Glycosignals in Cancer: Mechanisms of Malignant Phenotypes (Eds.: ), Springer Japan, Tokyo, 2016, pp. 141–161.
- 4
- 4aH. Paulsen, Č. Kolář, Angew. Chem. Int. Ed. Engl. 1978, 17, 771–771; Angew. Chem. 1978, 90, 823–824;
- 4bE. Y. Korchagina, I. M. Ryzhov, K. A. Byrgazov, I. S. Popova, S. i. N. Pokrovsky, N. V. Bovin, Mendeleev Commun. 2009, 19, 152–154;
- 4cW. Wang, T. Hu, P. A. Frantom, T. Zheng, B. Gerwe, D. S. Del Amo, S. Garret, R. D. Seidel III, P. Wu, Proc. Natl. Acad. Sci. USA 2009, 106, 16096–16101.
- 5J. Räbinä, J. Natunen, R. Niemelä, H. Salminen, K. Ilves, O. Aitio, H. Maaheimo, J. Helin, O. Renkonen, Carbohydr. Res. 1997, 305, 491–499.
- 6C. Zhao, Y. Wu, H. Yu, I. M. Shah, Y. Li, J. Zeng, B. Liu, D. A. Mills, X. Chen, Chem. Commun. 2016, 52, 3899–3902.
- 7L. A. Carpino, M. Ismail, G. A. Truran, E. M. E. Mansour, S. Iguchi, D. Ionescu, A. El-Faham, C. Riemer, R. Warrass, J. Org. Chem. 1999, 64, 4324–4338.
- 8Y. Li, M. Xue, X. Sheng, H. Yu, J. Zeng, V. Thon, Y. Chen, M. M. Muthana, P. G. Wang, X. Chen, Bioorg. Med. Chem. 2016, 24, 1696–1705.
- 9
- 9aI. A. Gagarinov, T. Fang, L. Liu, A. D. Srivastava, G. J. Boons, Org. Lett. 2015, 17, 928–931;
- 9bU. Ellervik, G. Magnusson, Carbohydr. Res. 1996, 280, 251–260.
- 10J. C. Castro-Palomino, G. Ritter, S. R. Fortunato, S. Reinhardt, L. J. Old, R. R. Schmidt, Angew. Chem. Int. Ed. Engl. 1997, 36, 1998–2001; Angew. Chem. 1997, 109, 2081–2085.
- 11R. Niemelä, J. Natunen, L. Penttilä, H. Salminen, J. Helin, H. Maaheimo, C. E. Costello, O. Renkonen, Glycobiology 1999, 9, 517–526.
- 12G. Sugiarto, K. Lau, J. Qu, Y. Li, S. Lim, S. Mu, J. B. Ames, A. J. Fisher, X. Chen, ACS Chem. Biol. 2012, 7, 1232–1240.
- 13O. Blixt, S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese, J. Stevens, N. Razi, D. J. Stevens, J. J. Skehel, I. van Die, D. R. Burton, I. A. Wilson, R. Cummings, N. Bovin, C. H. Wong, J. C. Paulson, Proc. Natl. Acad. Sci. USA 2004, 101, 17033–17038.
- 14T. B. H. Geijtenbeek, R. Torensma, S. J. van Vliet, G. C. F. van Duijnhoven, G. J. Adema, Y. van Kooyk, C. G. Figdor, Cell 2000, 100, 575–585.
- 15L. Nimrichter, M. M. Burdick, K. Aoki, W. Laroy, M. A. Fierro, S. A. Hudson, C. E. Von Seggern, R. J. Cotter, B. S. Bochner, M. Tiemeyer, K. Konstantopoulos, R. L. Schnaar, Blood 2008, 112, 3744–3752.
- 16M. Tiemeyer, S. J. Swiedler, M. Ishihara, M. Moreland, H. Schweingruber, P. Hirtzer, B. K. Brandley, Proc. Natl. Acad. Sci. USA 1991, 88, 1138–1142.
- 17
- 17aA. S. Gambaryan, A. B. Tuzikov, G. V. Pazynina, J. A. Desheva, N. V. Bovin, M. N. Matrosovich, A. I. Klimov, Virol. J. 2008, 5, 85;
- 17bZ. Wang, Z. S. Chinoy, S. G. Ambre, W. Peng, R. McBride, R. P. de Vries, J. Glushka, J. C. Paulson, G. J. Boons, Science 2013, 341, 379–383;
- 17cX. Xiong, A. Tuzikov, P. J. Coombs, S. R. Martin, P. A. Walker, S. J. Gamblin, N. Bovin, J. J. Skehel, Virus Res. 2013, 178, 12–14;
- 17dR. Xu, R. P. de Vries, X. Zhu, C. M. Nycholat, R. McBride, W. Yu, J. C. Paulson, I. A. Wilson, Science 2013, 342, 1230–1235.
- 18T. Hiono, M. Okamatsu, M. Igarashi, R. McBride, R. P. de Vries, W. Peng, J. C. Paulson, Y. Sakoda, H. Kida, Arch. Virol. 2016, 161, 307–316.
- 19W. Peng, K. M. Bouwman, R. McBride, O. C. Grant, R. J. Woods, M. H. Verheije, J. C. Paulson, R. P. de Vries, J. Virol. 2018, 92, e 0216-17.
- 20
- 20aS. Nishihara, H. Iwasaki, M. Kaneko, A. Tawada, M. Ito, H. Narimatsu FEBS Lett. 1999, 462, 289–294;
- 20bC. Dumon, E. Samain, B. Priem Biotechnol. Prog. 2004, 20, 412–419.