AlkAniline-Seq: Profiling of m7G and m3C RNA Modifications at Single Nucleotide Resolution
Virginie Marchand
Lorraine University, UMS2008 IBSLor CNRS-UL-INSERM, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorLilia Ayadi
Lorraine University, UMS2008 IBSLor CNRS-UL-INSERM, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Lorraine University, UMR7365 IMoPA CNRS-UL, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorFelix G. M. Ernst
RNA Molecular Biology, ULB-Cancer Research Center (U-CRC), Center for Microscopy and Molecular Imaging (CMMI), Fonds de la Recherche Scientifique (FRS), Université Libre de Bruxelles (ULB), BioPark campus, Gosselies, Belgium
Search for more papers by this authorJasmin Hertler
Institute of Pharmacy and Biochemistry, Johannes Gutenberg University Mainz, Staudingerweg 5, 55128 Mainz, Germany
Search for more papers by this authorValérie Bourguignon-Igel
Lorraine University, UMS2008 IBSLor CNRS-UL-INSERM, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Lorraine University, UMR7365 IMoPA CNRS-UL, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorAdeline Galvanin
Lorraine University, UMR7365 IMoPA CNRS-UL, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorAnnika Kotter
Institute of Pharmacy and Biochemistry, Johannes Gutenberg University Mainz, Staudingerweg 5, 55128 Mainz, Germany
Search for more papers by this authorMark Helm
Institute of Pharmacy and Biochemistry, Johannes Gutenberg University Mainz, Staudingerweg 5, 55128 Mainz, Germany
Search for more papers by this authorDenis L. J. Lafontaine
RNA Molecular Biology, ULB-Cancer Research Center (U-CRC), Center for Microscopy and Molecular Imaging (CMMI), Fonds de la Recherche Scientifique (FRS), Université Libre de Bruxelles (ULB), BioPark campus, Gosselies, Belgium
Search for more papers by this authorCorresponding Author
Yuri Motorin
Lorraine University, UMS2008 IBSLor CNRS-UL-INSERM, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Lorraine University, UMR7365 IMoPA CNRS-UL, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorVirginie Marchand
Lorraine University, UMS2008 IBSLor CNRS-UL-INSERM, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorLilia Ayadi
Lorraine University, UMS2008 IBSLor CNRS-UL-INSERM, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Lorraine University, UMR7365 IMoPA CNRS-UL, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorFelix G. M. Ernst
RNA Molecular Biology, ULB-Cancer Research Center (U-CRC), Center for Microscopy and Molecular Imaging (CMMI), Fonds de la Recherche Scientifique (FRS), Université Libre de Bruxelles (ULB), BioPark campus, Gosselies, Belgium
Search for more papers by this authorJasmin Hertler
Institute of Pharmacy and Biochemistry, Johannes Gutenberg University Mainz, Staudingerweg 5, 55128 Mainz, Germany
Search for more papers by this authorValérie Bourguignon-Igel
Lorraine University, UMS2008 IBSLor CNRS-UL-INSERM, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Lorraine University, UMR7365 IMoPA CNRS-UL, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorAdeline Galvanin
Lorraine University, UMR7365 IMoPA CNRS-UL, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorAnnika Kotter
Institute of Pharmacy and Biochemistry, Johannes Gutenberg University Mainz, Staudingerweg 5, 55128 Mainz, Germany
Search for more papers by this authorMark Helm
Institute of Pharmacy and Biochemistry, Johannes Gutenberg University Mainz, Staudingerweg 5, 55128 Mainz, Germany
Search for more papers by this authorDenis L. J. Lafontaine
RNA Molecular Biology, ULB-Cancer Research Center (U-CRC), Center for Microscopy and Molecular Imaging (CMMI), Fonds de la Recherche Scientifique (FRS), Université Libre de Bruxelles (ULB), BioPark campus, Gosselies, Belgium
Search for more papers by this authorCorresponding Author
Yuri Motorin
Lorraine University, UMS2008 IBSLor CNRS-UL-INSERM, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Lorraine University, UMR7365 IMoPA CNRS-UL, Biopôle UL, 9, Avenue de la Forêt de Haye, 54505 Vandoeuvre-les-Nancy, France
Search for more papers by this authorGraphical Abstract
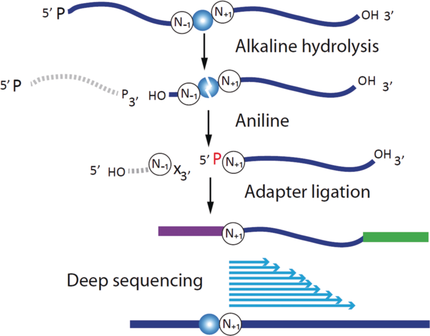
Deep sequencing was applied in a new concept of library preparation for detection of RNA modifications. Modified RNA is treated by alkaline hydrolysis, dephosphorylated, and subjected to aniline cleavage of abasic sites. The resulting 5′-phosphates in RNA are used for specific ligation of the sequencing adapter. The method can be applied for specific and sensitive detection of m7G, m3C, and (D) residues in RNAs.
Abstract
RNA modifications play essential roles in gene expression regulation. Only seven out of >150 known RNA modifications are detectable transcriptome-wide by deep sequencing. Here we describe a new principle of RNAseq library preparation, which relies on a chemistry based positive enrichment of reads in the resulting libraries, and therefore leads to unprecedented signal-to-noise ratios. The proposed approach eschews conventional RNA sequencing chemistry and rather exploits the generation of abasic sites and subsequent aniline cleavage. The newly generated 5′-phosphates are used as unique entry for ligation of an adapter in library preparation. This positive selection, embodied in the AlkAniline-Seq, enables a deep sequencing-based technology for the simultaneous detection of 7-methylguanosine (m7G) and 3-methylcytidine (m3C) in RNA at single nucleotide resolution. As a proof-of-concept, we used AlkAniline-Seq to comprehensively validate known m7G and m3C sites in bacterial, yeast, and human cytoplasmic and mitochondrial tRNAs and rRNAs, as well as for identifying previously unmapped positions.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie201810946-sup-0001-misc_information.pdf1.9 MB | Supplementary |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1D. Dominissini, Science 2014, 346, 1192.
- 2B. S. Zhao, I. A. Roundtree, C. He, Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42.
- 3Y. Saletore, K. Meyer, J. Korlach, I. D. Vilfan, S. Jaffrey, C. E. Mason, Genome Biol. 2012, 13, 175.
- 4D. Dominissini, S. Moshitch-Moshkovitz, M. Salmon-Divon, N. Amariglio, G. Rechavi, Nat. Protoc. 2013, 8, 176–189.
- 5D. Dominissini, S. Nachtergaele, S. Moshitch-Moshkovitz, E. Peer, N. Kol, M. S. Ben-Haim, Q. Dai, A. Di Segni, M. Salmon-Divon, W. C. Clark, et al., Nature 2016, 530, 441–446.
- 6T. M. Carlile, M. F. Rojas-Duran, B. Zinshteyn, H. Shin, K. M. Bartoli, W. V. Gilbert, Nature 2014, 515, 143–146.
- 7S. Schwartz, D. A. Bernstein, M. R. Mumbach, M. Jovanovic, R. H. Herbst, B. X. León-Ricardo, J. M. Engreitz, M. Guttman, R. Satija, E. S. Lander, et al., Cell 2014, 159, 148–162.
- 8J. E. Squires, H. R. Patel, M. Nousch, T. Sibbritt, D. T. Humphreys, B. J. Parker, C. M. Suter, T. Preiss, Nucleic Acids Res. 2012, 40, 5023–5033.
- 9B. Delatte, F. Wang, L. V. Ngoc, E. Collignon, E. Bonvin, R. Deplus, E. Calonne, B. Hassabi, P. Putmans, S. Awe, et al., Science 2016, 351, 282–285.
- 10U. Birkedal, M. Christensen-Dalsgaard, N. Krogh, R. Sabarinathan, J. Gorodkin, H. Nielsen, Angew. Chem. Int. Ed. 2015, 54, 451–455; Angew. Chem. 2015, 127, 461–465.
- 11E. M. Harcourt, A. M. Kietrys, E. T. Kool, Nature 2017, 541, 339–346.
- 12P. J. Hsu, H. Shi, C. He, Genome Biol. 2017, 18, 197.
- 13A. Visvanathan, K. Somasundaram, Bioessays 2018, https://doi.org/10.1002/bies.201700093.
- 14J. Choi, K.-W. Ieong, H. Demirci, J. Chen, A. Petrov, A. Prabhakar, S. E. O'Leary, D. Dominissini, G. Rechavi, S. M. Soltis, et al., Nat. Struct. Mol. Biol. 2016, 23, 110–115.
- 15J. Choi, G. Indrisiunaite, H. DeMirci, K.-W. Ieong, J. Wang, A. Petrov, A. Prabhakar, G. Rechavi, D. Dominissini, C. He, et al., Nat. Struct. Mol. Biol. 2018, 25, 208–216.
- 16X. Li, J. Peng, C. Yi, Methods Mol. Biol. 2017, 1562, 245–255.
- 17B. Molinie, J. Wang, K. S. Lim, R. Hillebrand, Z.-X. Lu, N. Van Wittenberghe, B. D. Howard, K. Daneshvar, A. C. Mullen, P. Dedon, et al., Nat. Methods 2016, 13, 692–698.
- 18M. Safra, A. Sas-Chen, R. Nir, R. Winkler, A. Nachshon, D. Bar-Yaacov, M. Erlacher, W. Rossmanith, N. Stern-Ginossar, S. Schwartz, Nature 2017, 551, 251–255.
- 19S. Hussain, J. Aleksic, S. Blanco, S. Dietmann, M. Frye, Genome Biol. 2013, 14, 215.
- 20M. Helm, Y. Motorin, Nat. Rev. Genet. 2017, 18, 275–291.
- 21S. Schwartz, Y. Motorin, RNA Biol. 2017, 14, 1124–1137.
- 22E. M. Novoa, C. E. Mason, J. S. Mattick, Nat. Rev. Mol. Cell Biol. 2017, 18, 339–340.
- 23V. S. Zueva, A. S. Mankin, A. A. Bogdanov, L. A. Baratova, Eur. J. Biochem. 1985, 146, 679–687.
- 24W. Wintermeyer, H. G. Zachau, FEBS Lett. 1975, 58, 306–309.
- 25P. A. Küpfer, C. J. Leumann, Nucleic Acids Res. 2007, 35, 58–68.
- 26R. Hauenschild, L. Tserovski, K. Schmid, K. Thüring, M.-L. Winz, S. Sharma, K.-D. Entian, L. Wacheul, D. L. J. Lafontaine, J. Anderson, et al., Nucleic Acids Res. 2015, 43, 9950–9964.
- 27C. J. Chetsanga, B. Bearie, C. Makaroff, Chem.-Biol. Interact. 1982, 41, 217–233.
- 28P. D. Lawley, P. Brookes, Biochem. J. 1963, 89, 127–138.
- 29C. J. Chetsanga, C. Makaroff, Chem.-Biol. Interact. 1982, 41, 235–249.
- 30J. White, Z. Li, R. Sardana, J. M. Bujnicki, E. M. Marcotte, A. W. Johnson, Mol. Cell. Biol. 2008, 28, 3151–3161.
- 31S. Figaro, L. Wacheul, S. Schillewaert, M. Graille, E. Huvelle, R. Mongeard, C. Zorbas, D. L. J. Lafontaine, V. Heurgué-Hamard, Mol. Cell. Biol. 2012, 32, 2254–2267.
- 32J. E. Smith, B. S. Cooperman, P. Mitchell, Biochemistry 1992, 31, 10825–10834.
- 33A. Alexandrov, M. R. Martzen, E. M. Phizicky, RNA 2002, 8, 1253–1266.
- 34P. Boccaletto, M. A. Machnicka, E. Purta, P. Piatkowski, B. Baginski, T. K. Wirecki, V. de Crécy-Lagard, R. Ross, P. A. Limbach, A. Kotter, et al., Nucleic Acids Res. 2018, 46, D 303–D307.
- 35S. D'Silva, S. J. Haider, E. M. Phizicky, RNA 2011, 17, 1100–1110.
- 36F. Xing, M. R. Martzen, E. M. Phizicky, RNA 2002, 8, 370–381.
- 37F. Xing, S. L. Hiley, T. R. Hughes, E. M. Phizicky, J. Biol. Chem. 2004, 279, 17850–17860.
- 38C. Zorbas, E. Nicolas, L. Wacheul, E. Huvelle, V. Heurgué-Hamard, D. L. J. Lafontaine, Mol. Biol. Cell 2015, 26, 2080–2095.
- 39J.-M. Chu, T.-T. Ye, C.-J. Ma, M.-D. Lan, T. Liu, B.-F. Yuan, Y.-Q. Feng, ACS Chem. Biol. 2018, https://doi.org/10.1021/acschembio.7b00906.
- 40N. Jonkhout, J. Tran, M. A. Smith, N. Schonrock, J. S. Mattick, E. M. Novoa, RNA 2017, 23, 1754–1769.
- 41L. P. Sarin, S. A. Leidel, RNA Biol. 2014, 11, 1555–1567.
- 42N. Husain, K. L. Tkaczuk, S. R. Tulsidas, K. H. Kaminska, S. Čubrilo, G. Maravić-Vlahoviček, J. M. Bujnicki, J. Sivaraman, Nucleic Acids Res. 2010, 38, 4120–4132.