Sensitive and Multiplexed On-chip microRNA Profiling in Oil-Isolated Hydrogel Chambers†
We acknowledge support from the NIH Center for Future Technologies in Cancer Care U54-EB-015403-01, the NCI grant 5R21A177393-02, Eni S.p.A. and NSF grant CMI-1120724.
Graphical Abstract
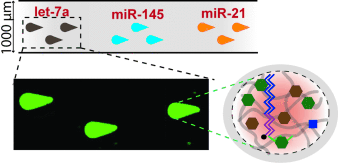
miRNA profiling: A versatile hydrogel-based microfluidic approach and novel amplification scheme were used for entirely on-chip, sensitive, and highly specific miRNA detection (let-7a, miR-145, and miR-21; see picture) without the risk of sequence bias. The approach uses photopolymerized hydrogel microposts for miRNA capture and labeling with a universal sequence. Fluorescence products are concentrated into the completely isolated gel posts.
Abstract
Although microRNAs (miRNAs) have been shown to be excellent indicators of disease state, current profiling platforms are insufficient for clinical translation. Here, we demonstrate a versatile hydrogel-based microfluidic approach and novel amplification scheme for entirely on-chip, sensitive, and highly specific miRNA detection without the risk of sequence bias. A simulation-driven approach is used to engineer the hydrogel geometry and the gel-reaction environment is chemically optimized for robust detection performance. The assay provides 22.6 fM sensitivity over a three log range, demonstrates multiplexing across at least four targets, and requires just 10.3 ng of total RNA input in a 2 hour and 15 minutes assay.