Constraining Cyclic Peptides To Mimic Protein Structure Motifs
Dr. Timothy A. Hill
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072 (Australia)
Search for more papers by this authorDr. Nicholas E. Shepherd
School of Molecular Biosciences, The University of Sydney, New South Wales 2006 (Australia)
Search for more papers by this authorDr. Frederik Diness
Center for Evolutionary Chemical Biology, Department of Chemistry, University of Copenhagen, Copenhagen (Denmark)
Search for more papers by this authorCorresponding Author
Prof. Dr. David P. Fairlie
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072 (Australia)
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072 (Australia)Search for more papers by this authorDr. Timothy A. Hill
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072 (Australia)
Search for more papers by this authorDr. Nicholas E. Shepherd
School of Molecular Biosciences, The University of Sydney, New South Wales 2006 (Australia)
Search for more papers by this authorDr. Frederik Diness
Center for Evolutionary Chemical Biology, Department of Chemistry, University of Copenhagen, Copenhagen (Denmark)
Search for more papers by this authorCorresponding Author
Prof. Dr. David P. Fairlie
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072 (Australia)
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072 (Australia)Search for more papers by this authorGraphical Abstract
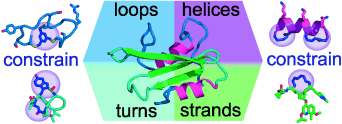
Short peptides can be constrained by cyclization to recreate key folded elements of protein structure, like β-strands and β-sheets, α-helices, and turn motifs. Coupled with internal molecular constraints, cyclization has led to many protease-resistant, potent and target-selective, biologically active compounds for use in biology and medicine.
Abstract
Many proteins exert their biological activities through small exposed surface regions called epitopes that are folded peptides of well-defined three-dimensional structures. Short synthetic peptide sequences corresponding to these bioactive protein surfaces do not form thermodynamically stable protein-like structures in water. However, short peptides can be induced to fold into protein-like bioactive conformations (strands, helices, turns) by cyclization, in conjunction with the use of other molecular constraints, that helps to fine-tune three-dimensional structure. Such constrained cyclic peptides can have protein-like biological activities and potencies, enabling their uses as biological probes and leads to therapeutics, diagnostics and vaccines. This Review highlights examples of cyclic peptides that mimic three-dimensional structures of strand, turn or helical segments of peptides and proteins, and identifies some additional restraints incorporated into natural product cyclic peptides and synthetic macrocyclic peptidomimetics that refine peptide structure and confer biological properties.
References
- 1
- 1ahttp://www.bmrb.wisc.edu/data_library/Diseases (BMRB entries relating to proteins and nucleic acids responsible for diseases of the human body);
- 1b Protein Structure and Function: From Sequence to Structure (Eds.: ), New Science Press, 2004, Chap. 1, pp. 2–49.
- 2M. R. Arkin, J. A. Wells, Nat. Rev. Drug Discovery 2004, 3, 301–317.
- 3
- 3aD. P. Fairlie, G. Abbenante, D. R. March, Curr. Med. Chem. 1995, 2, 654–686;
- 3bR. P. McGeary, D. P. Fairlie, Curr. Opin. Drug Discovery Dev. 1998, 1, 208–217;
- 3cD. P. Fairlie, M. L. West, A. K. Wong, Curr. Med. Chem. 1998, 5, 29–62.
- 4aA. Giannis, Angew. Chem. 1993, 105, 1303–1326; Angew. Chem. Int. Ed. Engl. 1993, 32, 1244–1267;
- 4bJ. Gante, Angew. Chem. 1994, 106, 1780–1802; Angew. Chem. Int. Ed. Engl. 1994, 33, 1699–1720;
- 4cJ. P. Schneider, J. W. Kelly, Chem. Rev. 1995, 95, 2169–2187;
- 4dE. M. Driggers, S. P. Hale, J. Lee, N. K. Terrett, Nat. Rev. Drug Discovery 2008, 7, 608–624;
- 4eJ. A. Robinson, Curr. Opin. Chem. Biol. 2011, 15, 379–386;
- 4fD. J. Craik, D. P. Fairlie, S. Liras, D. Price, Chem. Biol. Drug Des. 2013, 81, 136–147.
- 5
- 5aV. J. Hruby, F. al-Obeidi, W. Kazmierski, Biochem. J. 1990, 268, 249–262;
- 5bC. Toniolo, Int. J. Pept. Protein Res. 1990, 35, 287–300;
- 5cG. R. Marshall, Tetrahedron 1993, 49, 3547–3558;
- 5dR. M. Jones, P. D. Boatman, G. Semple, Y. J. Shin, S. Y. Tamura, Curr. Opin. Pharmacol. 2003, 3, 530–543;
- 5eS. Kee, S. D. S. Jois, Curr. Pharm. Des. 2003, 9, 1209–1224;
- 5fJ. D. A. Tyndall, B. Pfeiffer, G. Abbenante, D. P. Fairlie, Chem. Rev. 2005, 105, 793–826.
- 6
- 6aW. A. Loughlin, J. D. A. Tyndall, M. P. Glenn, D. P. Fairlie, Chem. Rev. 2004, 104, 6085–6117;
- 6bW. A. Loughlin, J. D. A. Tyndall, M. P. Glenn, T. A. Hill, D. P. Fairlie, Chem. Rev. 2010, 110( 6), PR32–PR69.
- 7
- 7aJ. S. Nowick, Acc. Chem. Res. 1999, 32, 287–296;
- 7bT. Moriuchi, T. Hirao, Chem. Soc. Rev. 2004, 33, 294–301;
- 7cJ. S. Nowick, Acc. Chem. Res. 2008, 41, 1319–1330;
- 7dP.-N. Cheng, J. D. Pham, J. S. Nowick, J. Am. Chem. Soc. 2013, 135, 5477–5492.
- 8
- 8aM. J. I. Andrews, A. B. Tabor, Tetrahedron 1999, 55, 11711–11743;
- 8bJ. W. Taylor, Biopolymers 2002, 66, 49–75;
- 8cM. J. Kelso, H. N. Hoang, W. N. Oliver, N. Sokolenko, D. R. March, T. G. Appleton, D. P. Fairlie, Angew. Chem. 2003, 115, 437–440;
10.1002/ange.200390096 Google ScholarAngew. Chem. Int. Ed. 2003, 42, 421–424;
- 8dN. E. Shepherd, G. Abbenante, D. P. Fairlie, Angew. Chem. 2004, 116, 2741–2744;
10.1002/ange.200352659 Google ScholarAngew. Chem. Int. Ed. 2004, 43, 2687–2690;
- 8eR. Fasan, R. L. A. Dias, K. Moehle, O. Zerbe, J. W. Vrijbloed, D. Obrecht, J. A. Robinson, Angew. Chem. 2004, 116, 2161–2164;
10.1002/ange.200353242 Google ScholarAngew. Chem. Int. Ed. 2004, 43, 2109–2112;
- 8fG. L. Verdine, G. J. Hilinski, Methods Enzymol. 2012, 503, 3–33;
- 8gY.-W. Kim, T. N. Grossmann, G. L. Verdine, Nat. Protoc. 2011, 6, 761–771;
- 8hG. H. Bird, W. C. Crannell, L. D. Walensky, Curr. Protoc. Chem. Biol. 2011, 3, 99–117;
- 8iA. B. Mahon, P. S. Arora, Drug Discovery Today Technol. 2012, 9, e 57–e62.
- 9
- 9aB. H. Zimm, J. K. Bragg, J. Chem. Phys. 1959, 31, 526–535;
- 9bJ. M. Scholtz, R. L. Baldwin, Annu. Rev. Biophys. Biomol. Struct. 1992, 21, 95–118;
- 9cH. J. Dyson, P. E. Wright, Annu. Rev. Biophys. Biophys. Chem. 1991, 20, 519–538.
- 10
- 10aE. J. Milner-White, Trends Pharmacol. Sci. 1989, 10, 70–74;
- 10bM. L. West, D. P. Fairlie, Trends Pharmacol. Sci. 1995, 16, 67–75;
- 10cO. H. Chan, B. H. Stewart, Drug Discovery Today 1996, 1, 461–473;
- 10dE. M. Topp, Med. Chem. Res. 1997, 7, 493–508;
- 10eK. A. Witt, T. J. Gillespie, J. D. Huber, R. D. Egleton, T. P. Davis, Peptides 2001, 22, 2329–2343;
- 10fH. J. Lee, Arch. Pharmacal Res. 2002, 25, 572–584.
- 11
- 11aC. A. Lipinski, F. Lombardo, B. W. Dominy, P. J. Feeney, Adv. Drug Delivery Rev. 1997, 23, 3–25;
- 11bC. A. Lipinski, J. Pharmacol. Toxicol. Methods 2000, 44, 235–249;
- 11cC. A. Lipinski, F. Lombardo, B. W. Dominy, P. J. Feeney, Adv. Drug Delivery Rev. 2001, 46, 3–26;
- 11dC. A. Lipinski, Drug Discovery Today Technol. 2004, 1, 337–341;
- 11eD. F. Veber, S. R. Johnson, H. Y. Cheng, B. R. Smith, K. W. Ward, K. D. Kopple, J. Med. Chem. 2002, 45, 2615–2623;
- 11fP. D. Leeson, B. Springthorpe, Nat. Rev. Drug Discovery 2007, 6, 881–890.
- 12
- 12aJ. D. A. Tyndall, D. P. Fairlie, J. Mol. Recognit. 1999, 12, 363–370;
10.1002/(SICI)1099-1352(199911/12)12:6<363::AID-JMR478>3.0.CO;2-M CAS PubMed Web of Science® Google Scholar
- 12bD. P. Fairlie, J. D. A. Tyndall, R. C. Reid, A. K. Wong, G. Abbenante, M. J. Scanlon, D. R. March, D. A. Bergman, C. L. C. Chai, B. A. Burkett, J. Med. Chem. 2000, 43, 1271–1281;
- 12cJ. D. A. Tyndall, T. Nall, D. P. Fairlie, Chem. Rev. 2005, 105, 973–1000;
- 12dP. K. Madala, J. D. A. Tyndall, T. Nall, D. P. Fairlie, Chem. Rev. 2010, 110( 6), PR1–PR31.
- 13R. S. Harrison, P. C. Sharpe, Y. Singh, D. P. Fairlie, Rev. Physiol. Biochem. Pharmacol. 2007, 159, 1–77.
- 14J. H. Brown, T. S. Jardetzky, J. C. Gorga, L. J. Stern, R. G. Urban, J. L. Strominger, D. C. Wiley, Nature 1993, 364, 33–39.
- 15Y. Qian, M. A. Blaskovich, M. Saleem, C. M. Seong, S. P. Wathen, A. D. Hamilton, S. M. Sebti, J. Biol. Chem. 1994, 269, 12410–12413.
- 16M. C. Hammond, B. Z. Harris, W. A. Lim, P. A. Bartlett, Chem. Biol. 2006, 13, 1247–1251.
- 17H. Yin, A. D. Hamilton, Angew. Chem. 2005, 117, 4200–4235; Angew. Chem. Int. Ed. 2005, 44, 4130–4163.
- 18
- 18aD. Leung, G. Abbenante, D. P. Fairlie, J. Med. Chem. 2000, 43, 305–341;
- 18bG. Abbenante, D. P. Fairlie, Med. Chem. 2005, 1, 71–104;
- 18c Protease inhibitors: Advances in Research and Application (Ed.: ), Scholarly Additions, Atlanta, 2012.
- 19R. C. Reid, M. J. Kelso, M. J. Scanlon, D. P. Fairlie, J. Am. Chem. Soc. 2002, 124, 5673–5683.
- 20
- 20aG. Abbenante, D. R. March, D. A. Bergman, P. A. Hunt, B. Garnham, R. J. Dancer, J. L. Martin, D. P. Fairlie, J. Am. Chem. Soc. 1995, 117, 10220–10226;
- 20bD. R. March, G. Abbenante, D. A. Bergman, R. I. Brinkworth, W. A. Wickramasinghe, J. Begun, J. L. Martin, D. P. Fairlie, J. Am. Chem. Soc. 1996, 118, 3375–3379;
- 20cR. C. Reid, L. K. Pattenden, J. D. A. Tyndall, J. L. Martin, T. Walsh, D. P. Fairlie, J. Med. Chem. 2004, 47, 1641–1651.
- 21
- 21aJ. D. A. Tyndall, R. C. Reid, D. P. Tyssen, D. K. Jardine, B. Todd, M. Passmore, D. R. March, L. K. Pattenden, D. Alewood, S. Hu, P. F. Alewood, C. J. Birch, J. L. Martin, D. P. Fairlie, J. Med. Chem. 2000, 43, 3495–3504;
- 21b“Mimicking extended conformations of protease substrates: designing cyclic peptidomimetics to inhibit HIV-1 Protease”: R. C. Reid, D. P. Fairlie in Advances in Amino Acid Mimetics and Peptidomimetics, Vol. 1 (Ed.: ), JAI, London, 1997, pp. 77–107.
10.1016/S1874-5113(97)80005-6 Google Scholar
- 22
- 22aR. C. Reid, D. R. March, M. Dooley, D. A. Bergman, G. Abbenante, D. P. Fairlie, J. Am. Chem. Soc. 1996, 118, 8511–8517;
- 22bJ. D. A. Tyndall, L. K. Pattenden, R. C. Reid, S. H. Hu, D. Alewood, P. F. Alewood, T. Walsh, D. P. Fairlie, J. L. Martin, Biochemistry 2008, 47, 3736–3744.
- 23J. D. A. Tyndall, D. P. Fairlie, Curr. Med. Chem. 2001, 8, 893–907.
- 24
- 24aS. Thaisrivongs, J. R. Blinn, D. T. Pals, S. R. Turner, J. Med. Chem. 1991, 34, 1276–1282;
- 24bA. E. Weber, T. A. Halgren, J. J. Doyle, R. J. Lynch, P. K. Siegl, W. H. Parsons, W. J. Greenlee, A. A. Patchett, J. Med. Chem. 1991, 34, 2692–2701;
- 24cA. E. Weber, M. G. Steiner, P. A. Krieter, A. E. Colletti, J. R. Tata, T. A. Halgren, R. G. Ball, J. J. Doyle, T. W. Schorn, R. A. Stearns, R. R. Miller, P. K. S. Siegl, W. J. Greenlee, A. A. Patchett, J. Med. Chem. 1992, 35, 3755–3773;
- 24dC. Sund, O. Belda, D. Wiktelius, C. Sahlberg, L. Vrang, S. Sedig, E. Hamelink, I. Henderson, T. Agback, K. Jansson, N. Borkakoti, D. Derbyshire, A. Eneroth, B. Samuelsson, Bioorg. Med. Chem. Lett. 2011, 21, 358–362.
- 25
- 25aA. K. Ghosh, T. Devasamudram, L. Hong, C. DeZutter, X. Xu, V. Weerasena, G. Koelsch, G. Bilcer, J. Tang, Bioorg. Med. Chem. Lett. 2005, 15, 15–20;
- 25bI. Rojo, J. A. Martin, H. Broughton, D. Timm, J. Erickson, H. C. Yang, J. R. McCarthy, Bioorg. Med. Chem. Lett. 2006, 16, 191–195;
- 25cS. J. Stachel, C. A. Coburn, S. Sankaranarayanan, E. A. Price, G. Wu, M. Crouthamel, B. L. Pietrak, Q. Huang, J. Lineberger, A. S. Espeseth, L. Jin, J. Ellis, M. K. Holloway, S. Munshi, T. Allison, D. Haxuda, A. J. Simon, S. L. Graham, J. P. Vacca, J. Med. Chem. 2006, 49, 6147–6150;
- 25dR. Machauer, S. Veenstra, J.-M. Rondeau, M. Tintelnot-Blomley, C. Betschart, U. Neumann, P. Paganetti, Bioorg. Med. Chem. Lett. 2009, 19, 1361–1365;
- 25eOther references listed in L. D. Pennington, D. A. Whittington, M. D. Bartberger, S. R. Jordan, H. Monenschein, T. T. Nguyen, B. H. Yang, Q. M. Xue, F. Vounatsos, R. C. Wahl, K. Chen, S. Wood, M. Citron, V. F. Patel, S. A. Hitchcock, W. Zhong, Bioorg. Med. Chem. Lett. 2013, 23, 4459–4464.
- 26
- 26aA. M. Silva, A. Y. Lee, S. V. Gulnik, P. Maier, J. Collins, T. N. Bhat, P. J. Collins, R. E. Cachau, K. E. Luker, I. Y. Gluzman, S. E. Francis, A. Oksman, D. E. Goldberg, J. W. Erickson, Proc. Natl. Acad. Sci. USA 1996, 93, 10034–10039;
- 26bK. Ersmark, M. Nervall, H. Gutierrez-de-Teran, E. Hamelink, L. K. Janka, J. C. Clemente, B. M. Dunn, A. Gogoll, B. Samuelsson, J. Aqvist, A. Hallberg, Bioorg. Med. Chem. 2006, 14, 2197–2208.
- 27
- 27aJ. H. Meyer, P. A. Bartlett, J. Am. Chem. Soc. 1998, 120, 4600–4609;
- 27bZ. Szewczuk, K. L. Rebholz, D. H. Rich, Int. J. Pept. Protein Res. 1992, 40, 233–242.
- 28
- 28aA. Marchetti, J. Ontoria, V. G. Matassa, Synlett 1999, 1000–1002;
- 28bY. S. Tsantrizos, Acc. Chem. Res. 2008, 41, 1252–1263;
- 28cY. S. Tsantrizos, Curr. Opin. Invest. Drugs 2009, 10, 871–881;
- 28dT.-I. Lin, O. Lenz, G. Fanning, T. Verbinnen, F. Delouvroy, A. Scholliers, K. Vermeiren, A. Rosenquist, M. Edlund, B. Samuelsson, L. Vrang, H. de Kock, P. Wigerinck, P. Raboisson, K. Simmen, Antimicrob. Agents Chemother. 2009, 53, 1377–1385;
- 28eJ. A. McCauley, C. J. McIntyre, M. T. Rudd, K. T. Nguyen, J. J. Romano, J. W. Butcher, K. F. Gilbert, K. J. Bush, M. K. Holloway, J. Swestock, B. L. Wan, S. S. Carroll, J. M. DiMuzio, D. J. Graham, S. W. Ludmerer, S. S. Mao, M. W. Stahlhut, C. M. Fandozzi, N. Trainor, D. B. Olsen, J. P. Vacca, N. J. Liverton, J. Med. Chem. 2010, 53, 2443–2463.
- 29F. Xue, C. T. Seto, J. Enzyme Inhib. Med. Chem. 2009, 24, 779–794.
- 30
- 30aB. E. Maryanoff, X. Qiu, K. P. Padmanabhan, A. Tulinsky, H. R., Jr. Almond, P. Andrade-Gordon, M. N. Greco, J. A. Kauffman, K. C. Nicolaou, A. Liu, P. H. Brungs, N. Fusetani, Proc. Natl. Acad. Sci. USA 1993, 90, 8048–8052;
- 30bV. Ganesh, A. Y. Lee, J. Clardy, A. Tulinsky, Protein Sci. 1996, 5, 825–835;
- 30c“Macrocyclic inhibitors of Serine Proteases”: M. N. Greco, B. E. Maryanoff in Advances in Amino Acid Mimetics and Peptidomimetics, Vol. 1 (Ed.: ), JAI, London, 1997, pp. 41–76.
10.1016/S1874-5113(97)80004-4 Google Scholar
- 31
- 31aL. J. MacPherson, E. K. Bayburt, M. P. Capparelli, R. S. Bohacek, F. H. Clarke, R. D. Ghai, Y. Sakane, C. J. Berry, J. V. Peppard, A. J. Trapani, J. Med. Chem. 1993, 36, 3821–3828;
- 31bC. B. Xue, X. He, J. Roderick, W. F. DeGrado, R. J. Cherney, K. D. Hardman, D. J. Nelson, R. A. Copeland, B. D. Jaffee, C. P. Decicco, J. Med. Chem. 1998, 41, 1745–1748.
- 32A. D. Abell, M. A. Jones, J. M. Coxon, J. D. Morton, S. G. Aitken, S. B. McNabb, H. Y. Lee, J. M. Mehrtens, N. A. Alexander, B. G. Stuart, A. T. Neffe, R. Bickerstaffe, Angew. Chem. 2009, 121, 1483–1486;
10.1002/ange.200805014 Google ScholarAngew. Chem. Int. Ed. 2009, 48, 1455–1458.
- 33
- 33aL. Gentilucci, A. Tolomelli, F. Squassabia, Curr. Med. Chem. 2006, 13, 2449–2466;
- 33bR. M. Jones, P. D. Boatman, G. Semple, Y.-J. Shin, S. Y. Tamura, Curr. Opin. Pharmacol. 2003, 3, 530–543.
- 34
- 34aC. J. White, A. K. Yudin, Nat. Chem. 2011, 3, 509–524;
- 34bR. J. Clark, D. J. Craik, Biopolymers 2010, 94, 414–422;
- 34cS. Jiang, Z. Li, K. Ding, P. P. Roller, Curr. Org. Chem. 2008, 12, 1502–1542;
- 34dJ. S. Davies, J. Pept. Sci. 2003, 9, 471–501;
- 34eM. Empting, O. Avrutina, R. Meusinger, S. Fabritz, M. Reinwarth, M. Biesalski, S. Voigt, G. Buntkowsky, H. Kolmar, Angew. Chem. 2011, 123, 5313–5317;
10.1002/ange.201008142 Google ScholarAngew. Chem. Int. Ed. 2011, 50, 5207–5211.
- 35
- 35aH. Kessler, B. Haase, Int. J. Pept. Protein Res. 1992, 39, 36–40;
- 35bM. Tamaki, S. Akabori, I. Muramatsu, J. Am. Chem. Soc. 1993, 115, 10492–10496.
- 36J. Chatterjee, D. Mierke, H. Kessler, J. Am. Chem. Soc. 2006, 128, 15164–15172.
- 37J. Chatterjee, D. F. Mierke, H. Kessler, Chem. Eur. J. 2008, 14, 1508–1517.
- 38A. I. Fernández-Llamazares, J. García, J. Adan, D. Meunier, F. Mitjans, J. Spengler, F. Albericio, Org. Lett. 2013, 15, 4572–4575.
- 39I. E. Valverde, F. Lecaille, G. Lalmanach, V. Aucagne, A. F. Delmas, Angew. Chem. 2012, 124, 742–746;
10.1002/ange.201107222 Google ScholarAngew. Chem. Int. Ed. 2012, 51, 718–722.
- 40
- 40aT. Rückle, P. de Lavallaz, M. Keller, P. Dumy, M. Mutter, Tetrahedron 1999, 55, 11281–11288;
- 40bD. Skropeta, K. A. Jolliffe, P. Turner, J. Org. Chem. 2004, 69, 8804–8809.
- 41L. S. Sonntag, S. Schweizer, C. Ochsenfeld, H. Wennemers, J. Am. Chem. Soc. 2006, 128, 14697–14703.
- 42P. Dumy, M. Keller, D. E. Ryan, B. Rohwedder, T. Wohr, M. Mutter, J. Am. Chem. Soc. 1997, 119, 918–925.
- 43G. Ruiz-Gómez, J. D. A. Tyndall, B. Pfeiffer, G. Abbenante, D. P. Fairlie, Chem. Rev. 2010, 110( 4), PR1–PR41.
- 44
- 44aA. K. Wong, A. M. Finch, G. K. Pierens, D. J. Craik, S. M. Taylor, D. P. Fairlie, J. Med. Chem. 1998, 41, 3417–3425;
- 44bA. M. Finch, A. K. Wong, N. J. Paczkowski, S. K. Wadi, D. J. Craik, D. P. Fairlie, S. M. Taylor, J. Med. Chem. 1999, 42, 1965–1974.
- 45D. R. March, L. M. Proctor, M. J. Stoermer, R. Sbaglia, G. Abbenante, R. C. Reid, T. M. Woodruff, K. Wadi, N. Paczkowski, J. D. A. Tyndall, S. M. Taylor, D. P. Fairlie, Mol. Pharmacol. 2004, 65, 868–879.
- 46“Discovery of Potent Antagonists of Human C5a Receptors”: S. M. Taylor, D. P. Fairlie in Structural Biology of Complement System (Ed.: ), CRC, Boca Raton, 2005, pp. 341–362.
10.1201/9780849350368.ch15 Google Scholar
- 47P. N. Monk, A. M. Scola, P. Madala, D. P. Fairlie, Br. J. Pharmacol. 2007, 152, 429–448.
- 48D. F. Veber, R. M. Freidinger, D. S. Perlow, W. J. Paleveda, F. W. Holly, R. G. Strachan, R. F. Nutt, B. H. Arison, C. Homnick, W. C. Randall, M. S. Glitzer, R. Saperstein, R. Hirschmann, Nature 1981, 292, 55–58.
- 49G. Melacini, Q. Zhu, M. Goodman, Biochemistry 1997, 36, 1233–1241.
- 50E. Pohl, A. Heine, G. M. Sheldrick, Z. Dauter, K. S. Wilson, J. Kallen, W. Huber, P. J. Pfaffli, Acta Crystallogr. Sect. D 1995, 51, 48–59.
- 51
- 51aA. Di Cianni, A. Carotenuto, D. Brancaccio, E. Novellino, J. C. Reubi, K. Beetschen, A. M. Papini, M. Ginanneschi, J. Med. Chem. 2010, 53, 6188–6197;
- 51bR. P. Cheng, D. J. Suich, H. Cheng, H. Roder, W. F. DeGrado, J. Am. Chem. Soc. 2001, 123, 12710–12711.
- 52
- 52aR. Thirumoorthy, J. R. Holder, R. M. Bauzo, N. G. J. Richards, A. S. Edison, C. Haskell-Luevano, J. Med. Chem. 2001, 44, 4114–4124;
- 52bA. Wilczynski, X. S. Wang, C. G. Joseph, Z. M. Xiang, R. M. Bauzo, J. W. Scott, N. B. Sorensen, A. M. Shaw, W. J. Millard, N. G. Richards, C. Haskell-Luevano, J. Med. Chem. 2004, 47, 2194–2207;
- 52cC. Thurieau, M. Feletou, P. Hennig, E. Raimbaud, E. Canet, J. L. Fauchere, J. Med. Chem. 1996, 39, 2095–2101.
- 53aG. Kotovych, J. R. Cann, J. M. Stewart, H. Yamamoto, Biochem. Cell Biol. 1998, 76, 257–266;
- 53bK. A. Carpenter, P. W. Schiller, R. Schmidt, B. C. Wilkes, Int. J. Pept. Protein Res. 1996, 48, 102–111;
- 53cW. M. Kazmierski, H. I. Yamamura, V. J. Hruby, J. Am. Chem. Soc. 1991, 113, 2275–2283.
- 54aM. Coles, V. Sowemimo, D. Scanlon, S. L. A. Munro, D. J. Craik, J. Med. Chem. 1993, 36, 2658–2665;
- 54bR. S. McDowell, K. A. Elias, M. S. Stanley, D. J. Burdick, J. P. Burnier, K. S. Chan, W. J. Fairbrother, R. G. Hammonds, G. S. Ingle, N. E. Jacobsen, D. L. Mortensen, T. E. Rawson, W. B. Won, R. G. Clark, T. C. Somers, Proc. Natl. Acad. Sci. USA 1995, 92, 11165–11169.
- 55aJ. Rizo, R. B. Sutton, J. Breslau, S. C. Koerber, J. Porter, A. T. Hagler, J. E. Rivier, L. M. Gierasch, J. Am. Chem. Soc. 1996, 118, 970–976;
- 55bP. Grieco, A. Lavecchia, M. Y. Cai, D. Trivedi, D. Weinberg, T. MacNeil, L. H. T. Van der Ploeg, V. J. Hruby, J. Med. Chem. 2002, 45, 5287–5294;
- 55cC. Fotsch, D. M. Smith, J. A. Adams, J. Cheetham, M. Croghan, E. M. Doherty, C. Hale, M. A. Jarosinski, M. G. Kelly, M. H. Norman, N. A. Tamayo, N. Xi, J. W. Baumgartner, Bioorg. Med. Chem. Lett. 2003, 13, 2337–2340.
- 56aM. A. Bednarek, T. Macneil, R. N. Kalyani, R. Tang, L. H. T. Van der Ploeg, D. H. Weinberg, Biochem. Biophys. Res. Commun. 1999, 261, 209–213;
- 56bM. A. Bednarek, T. MacNeil, R. N. Kalyani, R. Tang, L. H. T. Van der Ploeg, D. H. Weinberg, J. Med. Chem. 2001, 44, 3665–3672;
- 56cG. Skala, C. W. Smith, C. J. Taylor, J. H. Ludens, Science 1984, 226, 443–445.
- 57
- 57aW. Kowalczyk, D. Sobolewski, A. Prahl, I. Derdowska, A. Kwiatkowska, J. Slaninova, B. Lammek, J. Pept. Sci. 2006, 12, 181–182;
- 57bD. Sobolewski, A. Prahl, I. Derdowska, J. Slaninova, K. Kaczmarek, J. Zabrocki, B. Lammek, J. Pept. Sci. 2007, 13, 128–132.
- 58
- 58aR. Haubner, D. Finsinger, H. Kessler, Angew. Chem. Int. Ed. Engl. 1997, 36, 1375–1389;
- 58bR. Haubner, W. Schmitt, G. Holzemann, S. L. Goodman, A. Jonczyk, H. Kessler, J. Am. Chem. Soc. 1996, 118, 7881–7891;
- 58cS. L. Goodman, M. Picard, Trends Pharmacol. Sci. 2012, 33, 405–412;
- 58dC. Mas-Moruno, F. Rechenmacher, H. Kessler, Anti-Cancer Agents Med. Chem. 2010, 10, 753–768;
- 58eM. Schottelius, B. Laufer, H. Kessler, H. J. Wester, Acc. Chem. Res. 2009, 42, 969–980.
- 59
- 59aA. W. Tuin, D. K. Palachanis, A. Buizert, G. M. Grotenbreg, E. Spalburg, A. J. de Neeling, R. H. Mars-Groenendijk, D. Noort, G. A. van der Marel, H. S. Overkleeft, M. Overhand, Eur. J. Org. Chem. 2009, 4231–4241;
- 59bG. M. Grotenbreg, A. E. M. Buizert, A. L. Llamas-Saiz, E. Spalburg, P. A. V. van Hooft, A. J. de Neeling, D. Noort, M. J. van Raaij, G. A. van der Marel, H. S. Overkleeft, M. Overhand, J. Am. Chem. Soc. 2006, 128, 7559–7565.
- 60
- 60aC. Solanas, B. G. de La Torre, M. Fernandez-Reyes, C. M. Santiveri, M. A. Jimenez, L. Rivas, A. I. Jimenez, D. Andreu, C. Cativiela, J. Med. Chem. 2010, 53, 4119–4129;
- 60bM. Tamaki, I. Sasaki, M. Kokuno, M. Shindo, M. Kimura, Y. Uchida, Org. Biomol. Chem. 2010, 8, 1791–1797;
- 60cC. S. Dowd, S. Leavitt, G. Babcock, A. P. Godillot, D. Van Ryk, G. A. Canziani, J. Sodroski, E. Freire, I. M. Chaiken, Biochemistry 2002, 41, 7038–7046;
- 60dH. Tamamura, K. Hiramatsu, K. Miyamoto, A. Omagari, S. Oishi, H. Nakashima, N. Yamamoto, Y. Kuroda, T. Nakagawa, A. Otaka, N. Fujii, Bioorg. Med. Chem. Lett. 2002, 12, 923–928.
- 61H. N. Hoang, R. W. Driver, R. L. Beyer, A. K. Malde, G. T. Le, G. Abbenante, A. E. Mark, D. P. Fairlie, Angew. Chem. 2011, 123, 11303–11307; Angew. Chem. Int. Ed. 2011, 50, 11107–11111.
- 62
- 62aL. Belvisi, C. Gennari, A. Mielgo, D. Potenza, C. Scolastico, Eur. J. Org. Chem. 1999, 389–400;
10.1002/(SICI)1099-0690(199902)1999:2<389::AID-EJOC389>3.0.CO;2-7 CAS Web of Science® Google Scholar
- 62bM. K. Cho, S. S. Kim, M. R. Lee, J. Shin, J. Y. Lee, S. K. Lim, J. H. Baik, C. J. Yoon, I. Shin, W. Lee, J. Biochem. Mol. Biol. 2003, 36, 552–557;
- 62cS. Lindman, G. Lindeberg, F. Nyberg, A. Karlen, A. Hallberg, Bioorg. Med. Chem. 2000, 8, 2375–2383;
- 62dU. Rosenström, C. Sköld, B. Plouffe, H. Beaudry, G. Lindeberg, M. Botros, F. Nyberg, G. Wolf, A. Karlén, N. Gallo-Payet, A. Hallberg, J. Med. Chem. 2005, 48, 4009–4024;
- 62eZ. Q. Yuan, D. Blomberg, I. Sethson, K. Brickmann, K. Ekholm, B. Johansson, A. Nilsson, J. Kihlberg, J. Med. Chem. 2002, 45, 2512–2519;
- 62fK. Brickmann, Z. Yuan, I. Sethson, P. Somfai, J. Kihlberg, Chem. Eur. J. 1999, 5, 2241–2253.
10.1002/(SICI)1521-3765(19990802)5:8<2241::AID-CHEM2241>3.0.CO;2-L CAS Web of Science® Google Scholar
- 63
- 63aJ. A. Robinson, Acc. Chem. Res. 2008, 41, 1278–1288;
- 63bM. E. Pfeifer, K. Moehle, A. Linden, J. A. Robinson, Helv. Chim. Acta 2000, 83, 444–464;
10.1002/(SICI)1522-2675(20000216)83:2<444::AID-HLCA444>3.0.CO;2-R CAS Web of Science® Google Scholar
- 63cJ. Späth, F. Stuart, L. Y. Jiang, J. A. Robinson, Helv. Chim. Acta 1998, 81, 1726–1738.
- 64A. Gokhale, T. K. Weldeghiorghis, V. Taneja, S. D. Satyanarayanajois, J. Med. Chem. 2011, 54, 5307–5319.
- 65J. A. Robinson, S. C. Shankaramma, P. Jettera, U. Kienzl, R. A. Schwendener, J. W. Vrijbloed, D. Obrecht, Bioorg. Med. Chem. 2005, 13, 2055–2064.
- 66J. Liu, C. Li, S. Ke, S. D. Satyanarayanajois, J. Med. Chem. 2007, 50, 4038–4047.
- 67M. I. Garcia-Aranda, Y. Mirassou, B. Gautier, M. Martin-Martinez, N. Inguimbert, M. Vidal, M. T. Garcia-Lopez, M. A. Jimenez, R. Gonzalez-Muniz, M. J. P. de Vega, Bioorg. Med. Chem. 2011, 19, 7526–7533.
- 68J. H. Park, M. L. Waters, Org. Biomol. Chem. 2013, 11, 69–77.
- 69V. Celentano, D. Diana, L. De Rosa, C. Di Salvo, A. Romanelli, R. Fattorusso, L. D. D′Andrea, J. Pept. Sci. 2012, 18, S 173–S173.
- 70A. L. Jochim, P. S. Arora, ACS Chem. Biol. 2010, 5, 919–923.
- 71
- 71aK. Estieu-Gionnet, G. Guichard, Expert Opin. Drug Discovery 2011, 6, 937–963;
- 71bV. Azzarito, K. Long, N. S. Murphy, A. J. Wilson, Nat. Chem. 2013, 5, 161–173;
- 71cR. Dharanipragada, Future Med. Chem. 2013, 5, 831–849.
- 72R. S. Harrison, N. E. Shepherd, H. N. Hoang, G. Ruiz-Gomez, T. A. Hill, R. W. Driver, V. S. Desai, P. R. Young, G. Abbenante, D. P. Fairlie, Proc. Natl. Acad. Sci. USA 2010, 107, 11686–11691.
- 73
- 73aC. Bracken, J. Gulyas, J. W. Taylor, J. Baum, J. Am. Chem. Soc. 1994, 116, 6431–6432;
- 73bG. Osapay, J. W. Taylor, J. Am. Chem. Soc. 1990, 112, 6046–6051;
- 73cM. E. Houston, A. P. Campbell, B. Lix, C. M. Kay, B. D. Sykes, R. S. Hodges, Biochemistry 1996, 35, 10041–10050;
- 73dM. E. Houston, C. L. Gannon, C. M. Kay, R. S. Hodges, J. Pept. Sci. 1995, 1, 274–282.
- 74M. Bouvier, J. W. Taylor, J. Med. Chem. 1992, 35, 1145–1155.
- 75
- 75aS. Arttamangkul, T. F. Murray, G. E. Delander, J. V. Aldrich, J. Med. Chem. 1995, 38, 2410–2417;
- 75bF. D. T. Lung, N. Collins, D. Stropova, P. Davis, H. I. Yamamura, F. Porreca, V. J. Hruby, J. Med. Chem. 1996, 39, 1136–1141.
- 76M. Chorev, E. Roubini, R. L. McKee, S. W. Gibbons, M. E. Goldman, M. P. Caulfield, M. Rosenblatt, Biochemistry 1991, 30, 5968–5974.
- 77J. F. Hernandez, W. Kornreich, C. Rivier, A. Miranda, G. Yamamoto, J. Andrews, Y. Tache, W. Vale, J. Rivier, J. Med. Chem. 1993, 36, 2860–2867.
- 78K. A. Carpenter, R. Schmidt, S. Y. Yue, L. Hodzic, C. Pou, K. Payza, C. Godbout, W. Brown, E. Roberts, Biochemistry 1999, 38, 15295–15304.
- 79A. Kapurniotu, J. W. Taylor, J. Med. Chem. 1995, 38, 836–847.
- 80M. Zhang, B. Wu, H. Zhao, J. W. Taylor, J. Pept. Sci. 2002, 8, 125–136.
- 81M. Dong, J. A. Te, X. Xu, J. Wang, D. I. Pinon, L. Storjohann, A. J. Bordner, L. J. Miller, Biochemistry 2011, 50, 8181–8192.
- 82L. P. Miranda, K. A. Winters, C. V. Gegg, A. Patel, J. Aral, J. Long, J. Zhang, S. Diamond, M. Guido, S. Stanislaus, M. Ma, L. P. Li, M. J. Rose, L. Poppe, M. M. Veniant, J. Med. Chem. 2008, 51, 2758–2765.
- 83
- 83aT. R. Geistlinger, R. K. Guy, J. Am. Chem. Soc. 2001, 123, 1525–1526;
- 83bT. R. Geistlinger, R. K. Guy, J. Am. Chem. Soc. 2003, 125, 6852–6853;
- 83cT. R. Geistlinger, R. K. Guy, Methods Enzymol. 2003, 364, 223–246;
- 83dT. R. Geistlinger, A. C. McReynolds, R. K. Guy, Chem. Biol. 2004, 11, 273–281.
- 84N. E. Shepherd, H. N. Hoang, G. Abbenante, D. P. Fairlie, J. Am. Chem. Soc. 2005, 127, 2974–2983.
- 85N. E. Shepherd, H. N. Hoang, V. S. Desai, E. Letouze, P. R. Young, D. P. Fairlie, J. Am. Chem. Soc. 2006, 128, 13284–13289.
- 86N. E. Shepherd, R. S. Harrison, D. P. Fairlie, Curr. Drug Targets 2012, 13, 1348–1359.
- 87R. S. Harrison, G. Ruiz-Gomez, T. A. Hill, S. Y. Chow, N. E. Shepherd, R.-J. Lohman, G. Abbenante, H. N. Hoang, D. P. Fairlie, J. Med. Chem. 2010, 53, 8400–8408.
- 88T. Rao, G. Ruiz-Gomez, T. A. Hill, H. N. Hoang, D. P. Fairlie, J. M. Mason, PLoS One 2013, 8, e 59415.
- 89K. K. Khoo, M. J. Wilson, B. J. Smith, M.-M. Zhang, J. Gulyas, D. Yoshikami, J. E. Rivier, G. Bulaj, R. S. Norton, J. Med. Chem. 2011, 54, 7558–7566.
- 90A. Caporale, M. Sturlese, L. Gesiot, F. Zanta, A. Wittelsberger, C. Cabrele, J. Med. Chem. 2010, 53, 8072–8079.
- 91
- 91aH. E. Blackwell, R. H. Grubbs, Angew. Chem. 1998, 110, 3469–3472;
10.1002/(SICI)1521-3757(19981204)110:23<3469::AID-ANGE3469>3.0.CO;2-A Google ScholarAngew. Chem. Int. Ed. 1998, 37, 3281–3284;10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V CAS PubMed Web of Science® Google Scholar
- 91bH. E. Blackwell, J. D. Sadowsky, R. J. Howard, J. N. Sampson, J. A. Chao, W. E. Steinmetz, D. J. O’Leary, R. H. Grubbs, J. Org. Chem. 2001, 66, 5291–5302;
- 91cG. L. Verdine, G. J. Hilinski, Drug Discovery Today 2012, 9, e 41–e47.
- 92Y.-W. Kim, P. S. Kutchukian, G. L. Verdine, Org. Lett. 2010, 12, 3046–3049.
- 93Y.-W. Kim, G. L. Verdine, Bioorg. Med. Chem. Lett. 2009, 19, 2533–2536.
- 94C. E. Schafmeister, J. Po, G. L. Verdine, J. Am. Chem. Soc. 2000, 122, 5891–5892.
- 95D. J. Yeo, S. L. Warriner, A. J. Wilson, Chem. Commun. 2013, 49, 9131–9133.
- 96
- 96aL. D. Walensky, A. L. Kung, I. Escher, T. J. Malia, S. Barbuto, R. D. Wright, G. Wagner, G. L. Verdine, S. J. Korsmeyer, Science 2004, 305, 1466–1470;
- 96bL. D. Walensky, K. Pitter, J. Morash, K. J. Oh, S. Barbuto, J. Fisher, E. Smith, G. L. Verdine, S. J. Korsmeyer, Mol. Cell 2006, 24, 199–210.
- 97C. R. Braun, J. Mintseris, E. Gavathiotis, G. H. Bird, S. P. Gygi, L. D. Walensky, Chem. Biol. 2010, 17, 1325–1333.
- 98M. L. Stewart, E. Fire, A. E. Keating, L. D. Walensky, Nat. Chem. Biol. 2010, 6, 595–601.
- 99R. E. Moellering, M. Cornejo, T. N. Davis, B. C. Del, J. C. Aster, S. C. Blacklow, A. L. Kung, D. G. Gilliland, G. L. Verdine, J. E. Bradner, Nature 2009, 462, 182–188.
- 100
- 100aF. Bernal, A. F. Tyler, S. J. Korsmeyer, L. D. Walensky, G. L. Verdine, J. Am. Chem. Soc. 2007, 129, 2456–2457;
- 100bF. Bernal, M. Wade, M. Godes, T. N. Davis, D. G. Whitehead, A. L. Kung, G. M. Wahl, L. D. Walensky, Cancer Cell 2010, 18, 411–422.
- 101
- 101aC. J. Brown, S. T. Quah, J. Jong, A. M. Goh, P. C. Chiam, K. H. Khoo, M. L. Choong, M. A. Lee, L. Yurlova, K. Zolghadr, T. L. Joseph, C. S. Verma, D. P. Lane, ACS Chem. Biol. 2013, 8, 506–512;
- 101bS. J. Wei, T. Joseph, S. Chee, L. Li, L. Yurlova, K. Zolghadr, C. Brown, D. Lane, C. Verma, F. Ghadessy, PLoS One 2013, 8, e 81068.
- 102Y. S. Chang, B. Graves, V. Guerlavais, C. Tovar, K. Packman, K. H. To, K. A. Olson, K. Kesavan, P. Gangurde, A. Mukherjee, T. Baker, K. Darlak, C. Elkin, Z. Filipovic, F. Z. Qureshi, H. Cai, P. Berry, E. Feyfant, X. E. Shi, J. Horstick, D. A. Annis, A. M. Manning, N. Fotouhi, H. Nash, L. T. Vassilev, T. K. Sawyer, Proc. Natl. Acad. Sci. USA 2013, 110, E 3445–E3454.
- 103W. Kim, G. H. Bird, T. Neff, G. Guo, M. A. Kerenyi, L. D. Walensky, S. H. Orkin, Nat. Chem. Biol. 2013, 9, 643–650.
- 104C. Phillips, L. R. Roberts, M. Schade, R. Bazin, A. Bent, N. L. Davies, R. Moore, A. D. Pannifer, A. R. Pickford, S. H. Prior, C. M. Read, A. Scott, D. G. Brown, B. Xu, S. L. Irving, J. Am. Chem. Soc. 2011, 133, 9696–9699.
- 105
- 105aH. Zhang, F. Curreli, X. Zhang, S. Bhattacharya, A. A. Waheed, A. Cooper, D. Cowburn, E. O. Freed, A. K. Debnath, Retrovirology 2011, 8, 28–36;
- 105bG. H. Bird, N. Madani, A. F. Perry, A. M. Princiotto, J. G. Supko, X. He, E. Gavathiotis, J. G. Sodroski, L. D. Walensky, Proc. Natl. Acad. Sci. USA 2010, 107, 14093–14098; T.-L. Sun, Y. Sun, C.-C. Lee, H. W. Huang, Biophys. J. 2013, 104, 1923–1932.
- 106D. O. Sviridov, I. Z. Ikpot, J. Stonik, S. K. Drake, M. Amar, D. O. Osei-Hwedieh, G. Piszczek, S. Turner, A. T. Remaley, Biochem. Biophys. Res. Commun. 2011, 410, 446–451.
- 107N. N. Danial, L. D. Walensky, C.-Y. Zhang, C. S. Choi, J. K. Fisher, A. J. A. Molina, S. R. Datta, K. L. Pitter, G. H. Bird, J. D. Wikstrom, J. T. Deeney, K. Robertson, J. Morash, A. Kulkarni, S. Neschen, S. Kim, M. E. Greenberg, B. E. Corkey, O. S. Shirihai, G. I. Shulman, B. B. Lowell, S. J. Korsmeyer, Nat. Med. 2008, 14, 144–153.
- 108T. Okamoto, K. Zobel, A. Fedorova, C. Quan, H. Yang, W. J. Fairbrother, D. C. S. Huang, B. J. Smith, K. Deshayes, P. E. Czabotar, ACS Chem. Biol. 2013, 8, 297–302.
- 109G. H. Bird, E. Gavathiotis, J. L. Labelle, S. G. Katz, L. D. Walensky, ACS Chem. Biol. 2014, 9, 831–837.
- 110
- 110aS. Cantel, A. L. C. Isaad, M. Scrima, J. J. Levy, R. D. DiMarchi, P. Rovero, J. A. Halperin, A. M. D′Ursi, A. M. Papini, M. Chorev, J. Org. Chem. 2008, 73, 5663–5674;
- 110bM. Scrima, A. Le Chevalier-Isaad, P. Rovero, A. M. Papini, M. Chorev, A. M. D′Ursi, Eur. J. Org. Chem. 2010, 446–457.
- 111S. A. Kawamoto, A. Coleska, X. Ran, H. Yi, C.-Y. Yang, S. Wang, J. Med. Chem. 2012, 55, 1137–1146.
- 112
- 112aA. M. Leduc, J. O. Trent, J. L. Wittliff, K. S. Bramlett, S. L. Briggs, N. Y. Chirgadze, Y. Wang, T. P. Burris, A. F. Spatola, Proc. Natl. Acad. Sci. USA 2003, 100, 11273–11278;
- 112bA. K. Galande, K. S. Bramlett, T. P. Burris, J. L. Wittliff, A. F. Spatola, J. Pept. Res. 2004, 63, 297–302;
- 112cA. K. Galande, K. S. Bramlett, J. O. Trent, T. P. Burris, J. L. Wittliff, A. F. Spatola, ChemBioChem 2005, 6, 1991–1998.
- 113A. Muppidi, K. Doi, S. Edwardraja, E. J. Drake, A. M. Gulick, H.-G. Wang, Q. Lin, J. Am. Chem. Soc. 2012, 134, 14734–14737.
- 114A. Muppidi, Z. Wang, X. Li, J. Chen, Q. Lin, Chem. Commun. 2011, 47, 9396–9398.
- 115H. Jo, N. Meinhardt, Y. B. Wu, S. Kulkarni, X. Z. Hu, K. E. Low, P. L. Davies, W. F. DeGrado, D. C. Greenbaum, J. Am. Chem. Soc. 2012, 134, 17704–17713.
- 116A. M. Spokoyny, Y. Zou, J. J. Ling, H. Yu, Y.-S. Lin, B. L. Pentelute, J. Am. Chem. Soc. 2013, 135, 5946–5949.
- 117
- 117aM. M. Madden, V. C. I. Rivera, W. Song, Q. Lin, Chem. Commun. 2009, 5588–5590;
- 117bM. M. Madden, A. Muppidi, Z. Li, X. Li, J. Chen, Q. Lin, Bioorg. Med. Chem. Lett. 2011, 21, 1472–1475.
- 118
- 118aS. Kneissl, E. J. Loveridge, C. Williams, M. P. Crump, R. K. Allemann, ChemBioChem 2008, 9, 3046–3054;
- 118bP. Wysoczanski, R. J. R. J. Mart, E. J. Loveridge, C. Williams, S. B. M. Whittaker, M. P. Crump, R. K. Allemann, J. Am. Chem. Soc. 2012, 134, 7644–7647.
- 119A. D. de Araujo, H. N. Hoang, W. M. Kok, F. Diness, P. Gupta, T. A. Hill, D. A. Price, S. Liras, D. P. Fairlie, Angew. Chem. Int. Ed. 2014, 53, 6965–6999; Angew. Chem. 2014, 126, 7085–7089.
- 120D. Wang, K. Chen, G. Dimartino, P. S. Arora, Org. Biomol. Chem. 2006, 4, 4074–4081.
- 121
- 121aL. K. Henchey, A. L. Jochim, P. S. Arora, Curr. Opin. Chem. Biol. 2008, 12, 692–697;
- 121bA. Patgiri, A. L. Jochim, P. S. Arora, Acc. Chem. Res. 2008, 41, 1289–1300;
- 121cA. Patgiri, M. Z. Menzenski, A. B. Mahon, P. S. Arora, Nat. Protoc. 2010, 5, 1857–1865;
- 121dA. B. Mahon, P. S. Arora, Drug Discovery Today 2012, 9, e 57–e62.
- 122E. Cabezas, A. C. Satterthwait, J. Am. Chem. Soc. 1999, 121, 3862–3875.
- 123
- 123aR. N. Chapman, G. Dimartino, P. S. Arora, J. Am. Chem. Soc. 2004, 126, 12252–12253;
- 123bG. Dimartino, D. Y. Wang, R. N. Chapman, P. S. Arora, Org. Lett. 2005, 7, 2389–2392.
- 124A. Patgiri, M. R. Witten, P. S. Arora, Org. Biomol. Chem. 2010, 8, 1773–1776.
- 125
- 125aS. E. Miller, N. R. Kallenbach, P. S. Arora, Tetrahedron 2012, 68, 4434–4437;
- 125bA. B. Mahon, P. S. Arora, Chem. Commun. 2012, 48, 1416–1418.
- 126J. Liu, D. Wang, Q. Zheng, M. Lu, P. S. Arora, J. Am. Chem. Soc. 2008, 130, 4334–4337.
- 127L. K. Henchey, J. R. Porter, I. Ghosh, P. S. Arora, ChemBioChem 2010, 11, 2104–2107.
- 128D. Y. Wang, W. Liao, P. S. Arora, Angew. Chem. 2005, 117, 6683–6687; Angew. Chem. Int. Ed. 2005, 44, 6525–6529.
- 129D. Wang, M. Lu, P. S. Arora, Angew. Chem. 2008, 120, 1905–1908; Angew. Chem. Int. Ed. 2008, 47, 1879–1882.
- 130L. K. Henchey, S. Kushal, R. Dubey, R. N. Chapman, B. Z. Olenyuk, P. S. Arora, J. Am. Chem. Soc. 2010, 132, 941–943.
- 131A. Patgiri, K. K. Yadav, P. S. Arora, D. Bar-Sagi, Nat. Chem. Biol. 2011, 7, 585–587.
- 132
- 132aR. Fasan, R. L. Dias, K. Moehle, O. Zerbe, J. W. Vrijbloed, D. Obrecht, J. A. Robinson, Angew. Chem. 2004, 116, 2161–2164;
10.1002/ange.200353242 Google ScholarAngew. Chem. Int. Ed. 2004, 43, 2109–2112;
- 132bR. Fasan, R. L. Dias, K. Moehle, O. Zerbe, D. Obrecht, P. R. Mittl, M. G. Grütter, J. A. Robinson, ChemBioChem 2006, 7, 515–526.
- 133
- 133aK. Moehle, Z. Athanassiou, K. Patora, A. Davidson, G. Varani, J. A. Robinson, Angew. Chem. 2007, 119, 9260–9264;
10.1002/ange.200702801 Google ScholarAngew. Chem. Int. Ed. 2007, 46, 9101–9104;
- 133bA. Davidson, K. Patora-Komisarska, J. A. Robinson, G. Varani, Nucleic Acids Res. 2011, 39, 248–256.
- 134M. Seitz, P. Rusert, K. Moehle, A. Trkola, J. A. Robinson, Chem. Commun. 2010, 46, 7754–7756.
- 135
- 135aD. J. Craik, Science 2006, 311, 1563;
- 135bR. M. J. Liskamp, D. T. S. Rijkers, S. E. Bakker in Modern Supramolecular Chemistry: Strategies for Macrocycle Synthesis (Eds.: ), Wiley-VCH, Weinheim, 2008, pp. 1–26;
10.1002/9783527621484.ch1 Google Scholar
- 135cA. T. Bockus, C. M. McEwen, R. S. Lokey, Curr. Top. Med. Chem. 2013, 13, 821–836;
- 135d“Cyclic Peptides”: S. R. Adusumalli, A. K. Yudin, V. Rai in Natural Lactones and Lactams: Synthesis Occurrence and Biological Activity (Ed.: ), 2013, Wiley-VCH, Weinheim.
- 136
- 136aW. Xu, L. Li, L. Du, N. Tan, Acta Biochim. Biophys. Sin. 2011, 43, 757–762;
- 136bP. G. Arnison, M. J. Bibb, G. Bierbaum, A. A. Bowers, T. S. Bugni, G. Bulaj, J. A. Camarero, D. J. Campopiano, G. L. Challis, J. Clardy, P. D. Cotter, D. J. Craik, M. Dawson, E. Dittmann, S. Donadio, P. C. Dorrestein, K.-D. Entian, M. A. Fischbach, J. S. Garavelli, U. Goeransson, C. W. Gruber, D. H. Haft, T. K. Hemscheidt, C. Hertweck, C. Hill, A. R. Horswill, M. Jaspars, W. L. Kelly, J. P. Klinman, O. P. Kuipers, A. J. Link, W. Liu, M. A. Marahiel, D. A. Mitchell, G. N. Moll, B. S. Moore, R. Müller, S. K. Nair, I. F. Nes, G. E. Norris, B. M. Olivera, H. Onaka, M. L. Patchett, J. Piel, M. J. T. Reaney, S. Rebuffat, R. P. Ross, H.-G. Sahl, E. W. Schmidt, M. E. Selsted, K. Severinov, B. Shen, K. Sivonen, L. Smith, T. Stein, R. D. Süssmuth, J. R. Tagg, G.-L. Tang, A. W. Truman, J. C. Vederas, C. T. Walsh, J. D. Walton, S. C. Wenzel, J. M. Willey, W. A. van der Donk, Nat. Prod. Rep. 2013, 30, 108–160.
- 137
- 137aS. A. Sieber, M. A. Marahiel, Chem. Rev. 2005, 105, 715–738;
- 137bM. A. Fischbach, C. T. Walsh, Chem. Rev. 2006, 106, 3468–3496.
- 138
- 138aA. Rüegger, H. Kuhn, H. R. Lichti, R. Loosi, R. Huguenin, A. Quiquerez, A. von Wartburg, Helv. Chim. Acta 1976, 59, 1075–1092;
- 138bD. Faulds, K. L. Goa, P. Benfield, Drugs 1993, 45, 953–1040.
- 139T. Wieland, G. Luben, H. Ottenheym, J. Faesel, J. X. de Vries, W. Konz, A. Prox, J. Schmid, Angew. Chem. 1968, 80, 209–213;
10.1002/ange.19680800602 Google ScholarAngew. Chem. Int. Ed. Engl. 1968, 7, 204–208.
- 140
- 140aD. S. Dalisay, E. W. Rogers, A. S. Edison, T. F. Molinski, J. Nat. Prod. 2009, 72, 732–738;
- 140bD. S. Nielsen, H. N. Hoang, R.-J. Lohman, F. Diness, D. P. Fairlie, Org. Lett. 2012, 14, 5720–5723.
- 141
- 141aW. E. Houssen, M. Jaspars, ChemBioChem 2010, 11, 1803–1815;
- 141bD. Davyt, G. Serra, Mar. Drugs 2010, 8, 2755–2780;
- 141cR. A. Hughes, C. J. Moody, Angew. Chem. 2007, 119, 8076–8101;
10.1002/ange.200700728 Google ScholarAngew. Chem. Int. Ed. 2007, 46, 7930–7954.
- 142
- 142aE. Selva, G. Beretta, N. Montanini, G. S. Saddler, L. Gastaldo, P. Ferrari, R. Lorenzetti, P. Landini, F. Ripamonti, B. P. Goldstein, M. Berti, L. Montanaro, M. Denaro, J. Antibiot. 1991, 44, 693–701;
- 142bG. Heckmann, T. Bach, Angew. Chem. 2005, 117, 1223–1226;
10.1002/ange.200461715 Google ScholarAngew. Chem. Int. Ed. 2005, 44, 1199–1201;
- 142cK. C. Nicolaou, D. H. Dethe, G. Y. C. Leung, B. Zou, D. Y.-K. Chen, Chem. Asian J. 2008, 3, 413–429 and references cited therein.
- 143
- 143aK. Taori, V. J. Paul, H. Luesch, J. Am. Chem. Soc. 2008, 130, 1806–1807;
- 143bJ. Hong, H. Luesch, Nat. Prod. Rep. 2012, 29, 449–456.
- 144T. Velkov, P. E. Thompson, R. L. Nation, J. Li, J. Med. Chem. 2010, 53, 1898–1916.
- 145
- 145aS. Rudolph-Böhner, D. F. Mierke, L. Moroder, FEBS Lett. 1994, 349, 319–323;
- 145bL. Pearson, T. Mihali, M. Moffitt, R. Kellmann, B. Neilan, Mar. Drugs 2010, 8, 1650–1680;
- 145cH. Fujiki, M. Suganuma, Anti-Cancer Agents Med. Chem. 2011, 11, 4–18.
- 146
- 146aA. Liwo, A. Tempczyk, S. Ołdziej, M. D. Shenderovich, V. J. Hruby, S. Talluri, J. Ciarkowski, F. Kasprzykowski, L. Lankiewicz, Z. Grzonka, Biopolymers 1996, 38, 157–175;
10.1002/(SICI)1097-0282(199602)38:2<157::AID-BIP3>3.0.CO;2-U CAS PubMed Web of Science® Google Scholar
- 146bJ. P. Rose, C. K. Wu, C. D. Hsiao, E. Breslow, B. C. Wang, Nat. Struct Biol. 1996, 3, 163–169;
- 146cV. J. Hruby, Nat. Rev. Drug Discovery 2002, 1, 847–858.
- 147
- 147aH. Ueda, H. Nakajima, Y. Hori, T. Fujita, M. Nishimura, T. Goto, M. Okuhara, J. Antibiot. 1994, 47, 301–310;
- 147bH. Nakajima, Y. B. Kim, H. Terano, M. Yoshida, S. Horinouchi, Exp. Cell Res. 1998, 241, 126–133;
- 147cK. M. VanderMolen, W. McCulloch, C. J. Pearce, N. H. Oberlies, J. Antibiot. 2011, 64, 525–531.
- 148M. Góngora-Benítez, J. Tulla-Puche, F. Albericio, Chem. Rev. 2014, 114, 901–926.
- 149
- 149aJ. K. Klint, S. Senff, D. B. Rupasinghe, S. Y. Er, V. Herzig, G. M. Nicholson, G. F. King, Toxicon 2012, 60, 478–491;
- 149bG. Estrada, E. Villegas, G. Corzo, Nat. Prod. Rep. 2007, 24, 145–161.
- 150R. C. R. de La Vega, L. D. Possani, Toxicon 2005, 46, 831–844.
- 151Y. Yamazaki, T. Morita, Toxicon 2004, 44, 227–231.
- 152
- 152aB. G. Fry, K. Roelants, D. E. Champagne, H. Scheib, J. D. A. Tyndall, G. F. King, T. J. Nevalainen, J. A. Norman, R. J. Lewis, R. S. Norton, C. Renjifo, R. C. R. de la Vega, Annu. Rev. Genomics Hum. Genet. 2009, 10, 483–511;
- 152bN. R. Casewell, W. Wüster, F. J. Vonk, R. A. Harrison, B. G. Fry, Trends Ecol. Evolution 2013, 28, 219–229.
- 153S. Luckett, R. S. Garcia, J. J. Barker, A. V. Konarev, P. R. Shewry, A. R. Clarke, R. L. Brady, J. Mol. Biol. 1999, 290, 525–533.
- 154
- 154aH. Terlau, B. M. Olivera, Physiol. Rev. 2004, 84, 41–68;
- 154bB. M. Olivera, R. W. Teichert, Mol. Interventions 2007, 7, 251–260;
- 154cL. Azam, J. M. McIntosh, Acta Pharmacol. Sin. 2009, 30, 771–783;
- 154dR. Halai, D. J. Craik, Nat. Prod. Rep. 2009, 26, 526–536.
- 155R. J. Clark, H. Fischer, S. T. Nevin, D. J. Adams, D. J. Craik, J. Biol. Chem. 2006, 281, 23254–23263.
- 156A. Weinberg, G. Jin, S. Sieg, T. S. McCormick, Front. Immunol. 2012, 3, 1–9.
- 157
- 157aJ.-M. Schröder, J. Harder, Int. J. Biochem. Cell Biol. 1999, 31, 645–651;
- 157bM. V. Sawai, H. P. Jia, L. Liu, V. Aseyev, J. M. Wiencek, P. B. McCray, Jr., T. Ganz, W. R. Kearney, B. F. Tack, Biochemistry 2001, 40, 3810–3816.
- 158L. Thorstholm, D. J. Craik, Drug Discovery Today Technol. 2012, 9, e 13–e21.
- 159D. J. Craik, N. L. Daly, C. Waine, Toxicon 2001, 39, 43–60.
- 160
- 160aO. Saether, D. J. Craik, I. D. Campbel, K. Sletten, J. Juul, D. G. Normano, Biochemistry 1995, 34, 4147–4158;
- 160bS. T. Henriques, Y.-H. Huang, K. J. Rosengren, H. G. Franquelim, F. A. Carvalho, A. Johnson, S. Sonza, G. Tachedjian, M. A. R. B. Castanho, N. L. Daly, J. Biol. Chem. 2011, 286, 24231–24241.
- 161
- 161aL. A. Harris, M. D. Crowell, Curr. Opin. Mol. Ther. 2007, 9, 403–410;
- 161bN. Lee, A. Wald, Expert Opin. Drug Metab. Toxicol. 2011, 7, 651–659;
- 161cG. M. Pitari, Drug Des. Dev. Ther. 2013, 7, 351–360;
- 161dS. Sharma, T. Sharma, R. Dhingra, P. Tomar, S. Singh, M. Malhotra, T. R. Bhardwaj, Mini-Rev. Med. Chem. 2013, 13, 1685–1690.
- 162
- 162aT. Wieland, Peptides of Poisonous Amanita Mushrooms, Springer, New York, 1986, p. 256;
- 162bD. A. Bushnell, P. Cramer, R. D. Kornberg, Proc. Natl. Acad. Sci. USA 2002, 99, 1218–1222;
- 162cF. Enjalbert, S. Rapior, J. Nouguier-Soule, S. Guillon, N. Amouroux, C. Cabot, J. Toxicol. Clin. Toxicol. 2002, 40, 715–757.
- 163
- 163aF. Lynen, U. Wieland, Justus Liebigs Ann. Chem. 1938, 533, 93–117;
- 163bT. Wieland, Naturwissenschaften 1977, 64, 303–309.
- 164
- 164aS. D. Jolad, J. J. Hoffmann, S. J. Torrance, R. M. Wiedhopf, J. R. Cole, S. K. Arora, R. B. Bates, R. L. Gargiulo, G. R. Kriek, J. Am. Chem. Soc. 1977, 99, 8040–8044;
- 164bM. Zalacaín, E. Zaera, D. Vázquez, A. Jiménez, FEBS Lett. 1982, 148, 95–97.
- 165
- 165aT.-W. C. Leung, D. H. Williams, J. C. J. Barna, S. Foti, P. B. Oelrichs, Tetrahedron 1986, 42, 3333–3348;
- 165bS. D. Kahn, P. M. Booth, J. P. Waltho, D. H. Williams, J. Org. Chem. 1989, 54, 1901–1904.
- 166
- 166aC. Heinis, T. Rutherford, S. Freund, G. Winter, Nat. Chem. Biol. 2009, 5, 502–507;
- 166bA. Angelini, L. Cendron, S. Chen, J. Touati, G. Winter, G. Zanotti, C. Heinis, ACS Chem. Biol. 2012, 7, 817–821;
- 166cS. Chen, J. Morales-Sanfrutos, A. Angelini, B. Cutting, C. Heinis, ChemBioChem 2012, 13, 1032–1038;
- 166dV. Baeriswyl, C. Heinis, ChemMedChem 2013, 8, 377–384.
- 167W. Lian, P. Upadhyaya, C. A. Rhodes, Y. Liu, D. Pei, J. Am. Chem. Soc. 2013, 135, 11990–11995.
- 168J. S. Quartararo, P. Wu, J. A. Kritzer, ChemBioChem 2012, 13, 1490–1496.
- 169R. Roodbeen, B. Paaske, L. Jiang, J. K. Jensen, A. Christensen, J. T. Nielsen, M. Huang, F. A. Mulder, N. C. Nielsen, P. A. Andreasen, K. J. Jensen, ChemBioChem 2013, 14, 2179–2188.
- 170C. Chatterjee, M. Paul, L. Xie, W. A. van der Donk, Chem. Rev. 2005, 105, 633–683.
- 171D. L. Boger, Med. Res. Rev. 2001, 21, 356–381.
- 172J. R. Knox, R. F. Pratt, Antimicrob. Agents Chemother. 1990, 34, 1342–1347.