Are NMR-Derived Model Structures for β-Peptides Representative for the Ensemble of Structures Adopted in Solution?†
Alice Glättli
Laboratorium für Physikalische Chemie, ETH, ETH Hönggerberg, HCI, 8093 Zürich, Switzerland, Fax: (+41) 1-632-1039
Search for more papers by this authorWilfred F. van Gunsteren Prof.
Laboratorium für Physikalische Chemie, ETH, ETH Hönggerberg, HCI, 8093 Zürich, Switzerland, Fax: (+41) 1-632-1039
Search for more papers by this authorAlice Glättli
Laboratorium für Physikalische Chemie, ETH, ETH Hönggerberg, HCI, 8093 Zürich, Switzerland, Fax: (+41) 1-632-1039
Search for more papers by this authorWilfred F. van Gunsteren Prof.
Laboratorium für Physikalische Chemie, ETH, ETH Hönggerberg, HCI, 8093 Zürich, Switzerland, Fax: (+41) 1-632-1039
Search for more papers by this authorWe thank Prof. D. Seebach for challenging us to simulate the behavior of the peptide and Dr. K. Gademann for providing us with the 20 NMR model structures. Financial support from the Schweizer Nationalfonds (project number 2000-063590.00) and from the National Center of Competence in Research (NCCR) in Structural Biology of the Swiss National Science Foundation is gratefully acknowledged.
Graphical Abstract
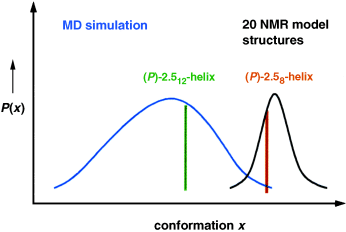
The data fit, but the structures are different. Two methods for the interpretation of NMR data of a β-hexapeptide are compared. While the simulated annealing procedure suggests the formation of a (P)-28-helix, unrestrained molecular dynamics simulations indicate that the NMR data can also be described by a much broader ensemble of conformations, in which (P)-2.512-helical structures are prominent (see diagram).
Supporting Information
Supporting information for this article is available on the WWW under http://www.wiley-vch.de/contents/jc_2002/2004/z460384_s.pdf or from the author.
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1O. Jardetzky, Biochim. Biophys. Acta 1980, 621, 227–232.
- 2J. Tropp, J. Chem. Phys. 1980, 76, 6035–6043.
- 3W. F. van Gunsteren, R. M. Brunne, P. Gros, R. C. van Schaik, C. A. Schiffer, A. E. Torda in Methods in Enzymology: Nuclear Magnetic Resonance, Vol. 239 (Eds.: ), Academic Press, New York, 1994, pp. 619–654.
- 4R. Abseher, S. Lüdemann, H. Schreiber, O. Steinhauser, J. Mol. Biol. 1995, 249, 604–624.
- 5A. M. J. J. Bonvin, A. T. Brünger, J. Biomol. NMR 1996, 7, 72–76.
- 6T. R. Schneider, A. T. Brünger, M. Nilges, J. Mol. Biol. 1999, 285, 727–740.
- 7C. A. E. M. Spronk, B. Sander, A. M. J. J. Bonvin, E. Krieger, G. W. Vuister, G. Vriend, J. Biomol. NMR 2003, 25, 225–234.
- 8A. E. Torda, W. F. van Gunsteren in Reviews in Computational Chemistry, Vol. III (Eds.: K. Lipkowitz, D. Boyd), VCH, New York, 1992, pp. 143–172.
- 9W. R. P. Scott, A. E. Mark, W. F. van Gunsteren, J. Biomol. NMR 1998, 12, 501–508.
- 10R. Abseher, S. Lüdemann, H. Schreiber, O. Steinhauser, J. Am. Chem. Soc. 1994, 116, 4006–4018.
- 11X. Daura, K. Gademann, B. Jaun, D. Seebach, W. F. van Gunsteren, A. E. Mark, Angew. Chem. 1999, 111, 249–253;
10.1002/(SICI)1521-3757(19990115)111:1/2<249::AID-ANGE249>3.0.CO;2-B Google ScholarAngew. Chem. Int. Ed. 1999, 38, 236–240.10.1002/(SICI)1521-3773(19990115)38:1/2<236::AID-ANIE236>3.0.CO;2-M CAS Web of Science® Google Scholar
- 12X. Daura, I. Antes, W. F. van Gunsteren, A. E. Mark, Proteins Struct. Funct. Genet. 1999, 36, 542–555.
10.1002/(SICI)1097-0134(19990901)36:4<542::AID-PROT17>3.0.CO;2-M CAS PubMed Web of Science® Google Scholar
- 13R. Bürgi, J. Pitera, W. F. van Gunsteren, J. Biomol. NMR 2001, 19, 305–320.
- 14C. Peter, X. Daura, W. F. van Gunsteren, J. Biomol. NMR 2001, 20, 297–310.
- 15A. Glättli, X. Daura, D. Seebach, W. F. van Gunsteren, J. Am. Chem. Soc. 2002, 124, 12 972–12 978.
- 16T. Hansson, C. Oostenbrink, W. F. van Gunsteren, Curr. Opin. Struct. Biol. 2002, 12, 190–196.
- 17K. Gademann, A. Häne, M. Rueping, B. Jaun, D. Seebach, Angew. Chem. 2003, 115, 1573–1575;
10.1002/ange.200250290 Google ScholarAngew. Chem. Int. Ed. 2003, 42, 1534–1537.
- 18D. Seebach, J. L. Matthews, Chem. Commun. 1997, 79, 2015–2022.
- 19R. P. Cheng, S. H. Gellman, W. F. DeGrado, Chem. Rev. 2001, 101, 3219–3232.
- 20D. Seebach, M. Overhand, F. N. M. Kühnle, B. Martinoni, L. Oberer, U. Hommel, H. Widmer, Helv. Chim. Acta 1996, 79, 913–941.
- 21D. Seebach, P. E. Ciceri, M. Overhand, B. Jaun, D. Rigo, L. Oberer, U. Hommel, H. Widmer, Helv. Chim. Acta 1996, 79, 2043–2066.
- 22D. H. Appella, L. A. Christianson, I. L. Karle, D. R. Powell, S. H. Gellman, J. Am. Chem. Soc. 1996, 118, 13 071–13 072.
- 23D. H. Appella, L. A. Christianson, D. A. Klein, D. R. Powell, X. Huang, J. J. Brachi, Jr.,S. H. Gellman, Nature 1997, 387, 381–384.
- 24D. Seebach, S. Abele, K. Gademann, G. Guichard, T. Hintermann, B. Jaun, J. L. Matthews, J. V. Schreiber, L. Oberer, U. Hommel, H. Widmer, Helv. Chim. Acta 1998, 81, 932–982.
- 25D. Seebach, S. Abele, K. Gademann, B. Jaun, Angew. Chem. 1999, 111, 1700–1703;
10.1002/(SICI)1521-3757(19990601)111:11<1700::AID-ANGE1700>3.0.CO;2-M Google ScholarAngew. Chem. Int. Ed. 1999, 38, 1595–1597.10.1002/(SICI)1521-3773(19990601)38:11<1595::AID-ANIE1595>3.0.CO;2-0 CAS PubMed Web of Science® Google Scholar
- 26W. F. van Gunsteren, S. R. Billeter, A. A. Eising, P. H. Hünenberger, P. Krüger, A. E. Mark, W. R. P. Scott, I. G. Tironi, Biomolecular Simulation: The GROMOS96 Manual and User Guide, vdf Hochschulverlag, ETH Zürich, Switzerland, 1996.
- 27W. R. P. Scott, P. H. Hünenberger, I. G. Tironi, A. E. Mark, S. R. Billeter, J. Fennen, A. E. Torda, T. Huber, P. Krüger, W. F. van Gunsteren, J. Phys. Chem. 1999, 103, 3596–3607.
- 28L. D. Schuler, X. Daura, W. F. van Gunsteren, J. Comput. Chem. 2001, 22, 1205–1218.
- 29A. T. Brünger, X-PLOR. A System for X-ray Crystallography and NMR, Yale University Press, New Haven, CT, USA, 1992.
- 30M. Karplus, J. Chem. Phys. 1959, 30, 11–15.
- 31X. Daura, W. F. van Gunsteren, A. E. Mark, Proteins Struct. Funct. Genet. 1999, 34, 269–280.
10.1002/(SICI)1097-0134(19990215)34:3<269::AID-PROT1>3.0.CO;2-3 CAS PubMed Web of Science® Google Scholar
- 32X. Daura, B. Jaun, D. Seebach, W. F. van Gunsteren, A. E. Mark, J. Mol. Biol. 1998, 280, 925–932.
- 33W. F. van Gunsteren, R. Bürgi, C. Peter, X. Daura, Angew. Chem. 2001, 113, 363–367;
10.1002/1521-3757(20010119)113:2<363::AID-ANGE363>3.0.CO;2-J Google ScholarAngew. Chem. Int. Ed. 2001, 40, 351–355.
- 34X. Daura, K. Gademann, H. Schäfer, B. Jaun, D. Seebach, W. F. van Gunsteren, J. Am. Chem. Soc. 2001, 123, 2393–2404.
- 35C. Peter, X. Daura, W. F. van Gunsteren, J. Am. Chem. Soc. 2000, 122, 7461–7466.
- 36R. Bürgi, X. Daura, A. E. Mark, M. Bellanda, B. Mammi, E. Peggion, W. F. van Gunsteren, J. Pept. Res. 2001, 57, 107–118.
- 37X. Daura, A. Glättli, P. Gee, C. Peter, W. F. van Gunsteren, Adv. Protein Chem. 2002, 62, 341–360.
- 38C. Peter, M. Rueping, H. J. Wörner, B. Jaun, D. Seebach, W. F. van Gunsteren, Chem. Eur. J. 2003, 9, 5838–5849.
- 39H. Yu, X. Daura, W. F. van Gunsteren, Proteins Struct. Funct. Genet. 2004, 54, 116–127.
- 40A. Pardi, M. Billeter, K. Wüthrich, J. Mol. Biol. 1984, 180, 741–751.
- 41A. de Marco, M. Llinás, K. Wüthrich, Biopolymers 1978, 17, 617–636.