3-Mercaptopropionate Dioxygenase (MDO)
Allison N Schmittou
Department of Chemistry & Biochemistry, University of Alabama, 250 Hackberry Lane, Tuscaloosa, AL, 35487 USA
Search for more papers by this authorYasmeen J Solano
Department of Physiology & Biophysics, University of California Irvine School of Medicine, 837 Health Sciences Road, Irvine, CA, 92617 USA
Search for more papers by this authorPhilip D Kiser
Department of Physiology & Biophysics, University of California Irvine School of Medicine, 837 Health Sciences Road, Irvine, CA, 92617 USA
Research Service, Louis Stokes Cleveland VA Medical Center, 10701 East Boulevard, Cleveland, OH, 44106 USA
Search for more papers by this authorBrad S Pierce
Department of Chemistry & Biochemistry, University of Alabama, 250 Hackberry Lane, Tuscaloosa, AL, 35487 USA
Search for more papers by this authorAllison N Schmittou
Department of Chemistry & Biochemistry, University of Alabama, 250 Hackberry Lane, Tuscaloosa, AL, 35487 USA
Search for more papers by this authorYasmeen J Solano
Department of Physiology & Biophysics, University of California Irvine School of Medicine, 837 Health Sciences Road, Irvine, CA, 92617 USA
Search for more papers by this authorPhilip D Kiser
Department of Physiology & Biophysics, University of California Irvine School of Medicine, 837 Health Sciences Road, Irvine, CA, 92617 USA
Research Service, Louis Stokes Cleveland VA Medical Center, 10701 East Boulevard, Cleveland, OH, 44106 USA
Search for more papers by this authorBrad S Pierce
Department of Chemistry & Biochemistry, University of Alabama, 250 Hackberry Lane, Tuscaloosa, AL, 35487 USA
Search for more papers by this authorAbstract
Bacterial 3-mercaptopropionate dioxygenase (MDO) is a mononuclear nonheme iron dioxygenase that catalyzes the O2-dependent oxidation of 3-mercaptopropioniate (3MPA) to produce 3-sulfinopropionate (3SPA). MDO is a member of the cupin superfamily of proteins and therefore shares a similar active site architecture as mammalian and bacterial cysteine dioxygenases (CDOs). However, unlike CDOs, which exhibit near exclusive specificity for l-cysteine (CYS), MDOs are more flexible with respect to accommodating other thiol-bearing substrates of comparable size to 3MPA. The active site of MDO and CDO share two major structural features. First, they both contain a mononuclear nonheme iron site coordinated by three histidine residues. Since all protein-derived ligands occupy one face of an octahedron, binding of dioxygen and organic substrate occurs at the opposite face of the 3-His facial triad. Second, both enzymes share a conserved sequence of outer Fe-coordination sphere amino acids (Ser, His, and Tyr) positioned adjacent to the iron site (∼ 4 Å). These residues participate in an extended proton relay network which ultimately donates an H-bond to the enzymatic Fe-site. To date, MDOs have only been identified among Gram-negative soil proteobacteria. It is believed that the high concentration of 3MPA present in coastal sediments can be attributed to bacterial catabolism of organosulfur compounds present in saline environments. Indirect evidence also suggests that 3MPA may play a more central role in bacterial sulfur metabolism than currently understood.
3D Structure
[This and all other 3D structural figures were generated using PyMOL (Schrödinger, Inc.).]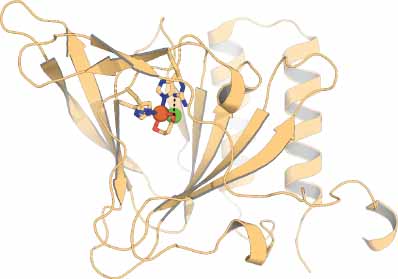
References
- 1NJ York, MM Lockart, S Sardar, N Khadka, W Shi, RE Stenkamp, J Zhang, PD Kiser and BS Pierce, J Biol Chem, 296, 1–19 (2021).
- 2BS Pierce, BP Subedi, S Sardar and JK Crowell, Biochemistry, 54, 7477–90 (2015).
- 3EP Tchesnokov, M Fellner, E Siakkou, T Kleffmann, LW Martin, S Aloi, IL Lamont, SM Wilbanks and GNL Jameson, J Biol Chem, 290, P24424–37 (2015).
- 4JK Crowell, S Sardar, MS Hossain, FW Foss and BS Pierce, Arch Biochem Biophys, 604, 86–94 (2016).
- 5SA Matthias Fellner, EP Tchesnokov and GNL Jameson, Biochemistry, 55, 1362–71 (2016).
- 6W Li and BS Pierce, Arch Biochem Biophys, 565, 49–56 (2015).
- 7C Doberstein, J Grote, JH Wübbeler and A Steinbüchel, J Biotechnol, 184, 187–98 (2014).
- 8N Bruland, JH Wübbeler and A Steinbüchel, J Biol Chem, 284, 660–72 (2008).
- 9M Stipanuk, C Simmons, PA Karplus and J Dominy, Amino Acids, 41, 91–102 (2010).
- 10RP Kiene and BF Taylor, Nature, 332, 148–50 (1988).
- 11J Zhang, F Wang, JD House and B Page, Limnol Oceanogr, 49, 2276–86 (2004).
- 12H Hu, SE Mylon and G Benoit, Limnol Oceanogr, 51, 2763–74 (2006).
- 13RP Kiene, KD Malloy and BF Taylor, Appl Environ Microbiol, 56, 156–61 (1990).
- 14KD Allen and RH White, FEMS Microbiol Lett, 363, fnw217 (2016).
- 15JH Wübbeler, N Bruland, M Wozniczka and A Steinbüchel, Microbiology, 156, 1221–33 (2010).
- 16RP Kiene and BF Taylor, Appl Environ Microbiol, 54, 2208–12 (1988).
- 17RP Kiene, LJ Linn and JA Bruton, J Sea Res, 43, 209–24 (2000).
- 18Y Zheng, J Wang, S Zhou, Y Zhang, J Liu, C-X Xue, BT Williams, X Zhao, L Zhao, X-Y Zhu, C Sun, H-H Zhang, T Xiao, G-P Yang, JD Todd and X-H Zhang, Nat Commun, 11, 4658 (2020).
- 19U Brandt, M Schürmann and A Steinbüchel, J Biol Chem, 289, 30800–9 (2014).
- 20JE Dominy, CR Simmons, LL Hirschberger, J Hwang, RM Coloso and MH Stipanuk, J Biol Chem, 282, 25189–98 (2007).
- 21L Ewetz and B Sorbo, Biochim Biophys Acta, 128, 296–305 (1966).
- 22B Sorbo and L Ewetz, Biochem Biophys Res Commun, 18, 359–63 (1965).
- 23JB Lombardini, TP Singer and PD Boyer, J Biol Chem, 244, 1172–5 (1969).
- 24C Gordon, P Emery, H Bradley and H Waring, Lancet, 229, 25–6 (1992).
- 25JE Dominy Jr CR Simmons, PA Karplus, AM Gehring and MH Stipanuk, J Bacteriol, 188, 5561–9 (2006).
- 26MT Heafield, S Fearn, GB Steventon, RH Waring, AC Williams and SG Sturman, Neurosci Lett, 110, 216–20 (1990).
- 27B Sarkar, M Kulharia and AK Mantha, Int J Exp Pathol, 98, 52–66 (2017).
- 28CD Brown, ML Neidig, MB Neibergall, JD Lipscomb and EI Solomon, J Am Chem Soc, 129, 7427–38 (2007).
- 29K Yamaguchi, S Sakakibara, K Koga and I Ueda, Biochim Biophys Acta, 237, 502–12 (1971).
- 30TL Perry, MG Norman, VW Yong, S Whiting, JU Crichton, S Hansen and SJ Kish, Ann Neurol, 18, 482–9 (1985).
- 31H Bradley, A Gough, RS Sokhi, A Hassell, R Waring and P Emery, J Rheumatol, 21, 1192–6 (1994).
- 32R Deth, C Muratore, J Benzecry, V-A Power-Charnitsky and M Waly, Neurotoxicology, 29, 190–201 (2008).
- 33SJ James, P Cutler, S Melnyk, S Jernigan, L Janak, DW Gaylor and JA Neubrander, Am J Clin Nutr, 80, 1611–7 (2004).
- 34KG Reddie and KS Carroll, Curr Opin Chem Biol, 12, 746–54 (2008).
- 35PG Winyard, CJ Moody and C Jacob, Trends Biochem Sci, 30, 453–61 (2005).
- 36D Trachootham, J Alexandre and P Huang, Nat Rev Drug Discov, 8, 579–91 (2009).
- 37DP Behave, WB Muse and KS Carroll, Infect Disord Drug Targets, 7, 140–58 (2007).
- 38DM Gunawardana, KC Heathcote and E Flashman, FEBS J, 289, 5426–39 (2021).
- 39DA Weits, B Giuntoli, M Kosmacz, S Parlanti, H-M Hubberten, H Riegler, R Hoefgen, P Perata, JT van Dongen and F Licausi, Nat Commun, 5, 3425 (2014).
- 40MD White, M Klecker, RJ Hopkinson, DA Weits, C Mueller, C Naumann, R O'Neill, J Wickens, J Yang, JC Brooks-Bartlett, EF Garman, TN Grossmann, N Dissmeyer and E Flashman, Nat Commun, 8, 14690 (2017).
- 41N Masson, TP Keeley, B Giuntoli, MD White, ML Puerta, P Perata, RJ Hopkinson, E Flashman, F Licausi and PJ Ratcliffe, Science, 365, 65–9 (2019).
- 42RL Fernandez, LD Elmendorf, RW Smith, CA Bingman, BG Fox and TC Brunold, Biochemistry, 60, 3728–37 (2021).
- 43Y Wang, I Shin, J Li and A Liu, J Biol Chem, 297, 101176 (2021).
- 44S Aloi, CG Davies, PA Karplus, SM Willbanks and GNL Jameson, Biochemistry, 58, 2398–407 (2019).
- 45S Khuri, FT Bakker and JM Dunwell, Mol Biol Evol, 18, 593–605 (2001).
- 46J Pei, BH Kim and NV Grishin, Nucleic Acids Res, 36, 2295–300 (2008).
- 47S Sardar, A Weitz, MP Hendrich and BS Pierce, Biochemistry, 58, 5135–50 (2019).
- 48NJ York, MM Lockart and BS Pierce, Inorg Chem, 60, 18639–51 (2021).
- 49CM Driggers, SJ Hartman and PA Karplus, Protein Sci, 24, 154–61 (2015).
- 50S Guindon, JF Dufayard, V Lefort, M Anisimova, W Hordijk and O Gascuel, Syst Biol, 59, 307–21 (2010).
- 51V Lefort, JE Longueville and O Gascuel, Mol Biol Evol, 34, 2422–4 (2017).
- 52BS Pierce, JD Gardner, LJ Bailey, TC Brunold and BG Fox, Biochemistry, 46, 8569–78 (2007).
- 53LL Stookey, Anal Chem, 42, 779–81 (1970).
- 54JD Gardner, BS Pierce, BG Fox and TC Brunold, Biochemistry, 49, 6033–41 (2010).
- 55JK Crowell, W Li and BS Pierce, Biochemistry, 53, 7541–8 (2014).
- 56M Fellner, LM Doughty, GNL Jameson and SM Willbanks, Anal Biochem, 459, 56–60 (2014).
- 57JR Winther and C Thorpe, Biochim Biophys Acta, 1840, 838–46 (2014).
- 58GA Bagiyan, IK Koroleva, NV Soroka and AV Ufimtsev, Russ Chem Bull, 52, 1135–41 (2003).
- 59K Ulrich and U Jakob, Free Radic Biol Med, 140, 14–27 (2019).
- 60C Chu, D Stamatelatos and K McNeill, Environ Sci Process Impacts, 19, 1518–27 (2017).
- 61JA Crawford, W Li and BS Pierce, Biochemistry, 50, 10241–53 (2011).
- 62E Siakkou, SM Wilbanks and GNL Jameson, Anal Biochem, 405, 127–31 (2010).
- 63P Salgado, T Visnevschi-Necrasov, RP Kiene, I Azevedo, ACS Rocha, CMR Almeida and C Magalhães, J Chromatogr B, 992, 103–8 (2015).
- 64GL Newton and RC Fahey, in L Packer (ed.), Biothiols Part A Monothiols and Dithiols, Protein Thiols, and Thiyl Radicals, Methods in Enzymology, Academic Press, San Diego, New York, Boston, London, Sydney, Tokyo, Toronto, pp 148–66 (1995).
- 65W Li, EJ Blaesi, MD Pecore, JK Crowell and BS Pierce, Biochemistry, 52, 9104–19 (2013).
- 66MT Rutledge, E Siakkou, SM Willbanks and GNL Jameson, Biochim Biophys Acta, 1814, 2003–9 (2011).
- 67P Panuwet, RE Hunter Jr PE D'Souza, X Chen, SA Radford, JR Cohen, ME Marder, K Kartavenka, PB Ryan and DB Barr, Crit Rev Anal Chem, 46, 93–105 (2016).
- 68R Banerjee and JD Lipscomb, Acc Chem Res, 54, 2185–95 (2021).
- 69CCG Yeh, C Pierides, GNL Jameson and SP de Visser, Chem Eur J, 27, 13793–806 (2021).
- 70CM Driggers, KM Kean, LL Hirschberger, RB Cooley, MH Stipanuk and PA Karplus, J Mol Biol, 428, 3999–4012 (2016).
- 71M Fellner, E Siakkou, AS Faponle, EP Tchesnokov, SP de Visser, SM Wilbanks and GN Jameson, J Biol Inorg Chem, 21, 501–10 (2016).
- 72CR Simmons, Q Liu, Q Huang, Q Hao, TP Begley, PA Karplus and MH Stipanuk, J Biol Chem, 281, 18723–33 (2006).
- 73CG Davies, M Fellner, EP Tchesnokov, SM Wilbanks and GNL Jameson, Biochemistry, 53, 7961–8 (2014).
- 74EJ Blaesi, BG Fox and TC Brunold, Biochemistry, 54, 2874–84 (2015).
- 75S Ye, W Xa, L Wei, D Tang, P Sun, M Bartlam and Z Rao, J Biol Chem, 282, 3391–402 (2007).
- 76JE Dominy, J Hwang, S Guo, LL Hirschberger, S Zhang and MH Stipanuk, J Biol Chem, 283, 12188–201 (2008).
- 77CW Njeri and HR Ellis, Arch Biochem Biophys, 558, 61–9 (2014).
- 78J Li, T Koto, I Davis and A Liu, Biochemistry, 58, 2218–27 (2019).
- 79G Dodson and A Wlodawer, Trends Biochem Sci, 23, 347–52 (1998).
- 80DT Petasis and MP Hendrich, Methods Enzymol, 563, 171–208 (2015).
- 81MP Hendrich and PG Debrunner, Biophys J, 56, 489–506 (1989).
- 82AC Weitz, N Giri, JD Caranto, DM Kurtz, EL Bominaar and MP Hendrich, J Am Chem Soc, 139, 12009–19 (2017).
- 83DL Tierney, AM Rocklin, JD Lipscomb, L Que and BM Hoffman, J Am Chem Soc, 127, 7005–13 (2005).
- 84TM Casey, PK Grzyska, RP Hausinger and J McCracken, J Phys Chem B, 117, 10384–94 (2013).
- 85J McCracken, BE Eser, D Mannikko, MD Krzyaniak and PF Fitzpatrick, Biochemistry, 54, 3759–71 (2015).
- 86AC McQuilken, Y Ha, KD Sutherlin, MA Siegler, KO Hodgson, B Hedman, EI Solomon, GNL Jameson and DP Goldberg, J Am Chem Soc, 135, 14024–7 (2013).
- 87AR Diebold, ML Neidig, GR Moran, GD Straganz and EI Solomon, Biochemistry, 49, 6945–52 (2010).
- 88A Wanat, T Schneppensieper, G Stochel, R van Eldik, E Bill and K Wieghardt, Inorg Chem, 41, 4–10 (2002).
- 89M Li, D Bonnet, E Bill, F Neese, T Weyhermèuller, N Blum, D Sellmann and K Wieghardt, Inorg Chem, 41, 3444–56 (2002).
- 90D Sellmann, N Blum, FW Heinemann and BA Hess, Chem Eur J, 7, 1874–80 (2001).
10.1002/1521-3765(20010504)7:9<1874::AID-CHEM1874>3.0.CO;2-2 CAS PubMed Web of Science® Google Scholar
- 91J McCracken, TM Casey and RP Hausinger, Appl Magn Reson (2020).
- 92RJ Martinie, J Livada, W-C Chang, MT Green, C Krebs, JM Bollinger and A Silakov, J Am Chem Soc, 137, 6912–9 (2015).
- 93AJ Mitchell, NP Dunham, RJ Martinie, JA Bergman, CJ Pollock, K Hu, BD Allen, W-C Chang, A Silakov, JM Bollinger Jr C Krebs and AK Boal, J Am Chem Soc, 139, 13830–6 (2017).
- 94C-P Yu, Y Tang, L Cha, S Milikisiyants, TI Smirnova, AI Smirnov, Y Guo and W-c Chang, J Am Chem Soc, 140, 15190–3 (2018).
- 95DM Arciero, JD Lipscomb, BH Huynh, TA Kent and E Münck, J Biol Chem, 258, 14981–91 (1983).
- 96ES Traore and A Liu, ACS Catal, 12, 6191–208 (2022).
- 97JB Gordon, JP McGale, JR Prendergast, Z Shirani-Sarmazeh, MA Siegler, GNL Jameson and DP Goldberg, J Am Chem Soc, 140, 14807–22 (2018).
- 98EP Tchesnokov, AS Faponle, CG Davies, MG Quesne, R Turner, M Fellner, RJ Souness, SM Wilbanks, SP de Visser and GNL Jameson, Chem Commun (Camb), 52, 8814–7 (2016).
- 99S Aluri and SP de Visser, J Am Chem Soc, 129, 14846–7 (2007).
- 100SP de Visser and GD Straganz, J Phys Chem A, 113, 1835–46 (2009).
- 101D Kumar, W Thiel and SP de Visser, J Am Chem Soc, 133, 3869–82 (2011).
- 102CR Simmons, K Krishnamoorthy, SL Granett, DJ Schuller, JE Dominy Jr TP Begley, MH Stipanuk and PA Karplus, Biochemistry, 47, 11390–2 (2008).
- 103EI Solomon, S Goudarzi and KD Sutherlin, Biochemistry, 55, 6363–74 (2016).
- 104JC Price, EW Barr, B Tirupati, JM Bollinger and C Krebs, Biochemistry, 42, 7497–508 (2003).
- 105LM Hoffart, EW Barr, RB Guyer, JM Bollinger and C Krebs, Proc Natl Acad Sci USA, 103, 14738–43 (2006).
- 106D Galonic Fujimori, EW Barr, ML Matthews, GM Koch, JR Yonce, CT Walsh, JM Bollinger, C Krebs and PJ Riggs-Gelasco, J Am Chem Soc, 129, 13408–9 (2007).
- 107SJ Lange, H Miyake and L Que, J Am Chem Soc, 121, 6330–1 (1999).
- 108MH Lim, J-U Rohde, A Stubna, MR Bukowski, M Costas, RYN Ho, E Münck, W Nam and L Que, Proc Natl Acad Sci USA, 100, 3665–70 (2003).
- 109LA Tyler, JC Noveron, MM Olmstead and PK Mascharak, Inorg Chem, 38, 616–7 (1999).
- 110I Kung, D Schweitzer, J Shearer, WD Taylor, HL Jackson, S Lovell and JA Kovacs, J Am Chem Soc, 122, 8299–300 (2000).
- 111RM Buonomo, I Font, MJ Maguire, JH Reibenspies, T Tuntulani and MY Darensbourg, J Am Chem Soc, 117, 963–73 (1995).
- 112A Dey, M Chow, K Taniguchi, P Lugo-Mas, S Davin, M Maeda, JA Kovacs, M Odaka, KO Hodgson, B Hedman and EI Solomon, J Am Chem Soc, 128, 533–41 (2005).
- 113CM Driggers, RB Cooley, B Sankaran, LL Hirschberger, MH Stipanuk and PA Karplus, J Mol Biol, 425, 3121–36 (2013).
- 114RJ Souness, T Kleffmann, EP Tchesnokov, SM Wilbanks, GB Jameson and GNL Jameson, Biochemistry, 52, 7606–17 (2013).



