The European Badger (Meles meles) as a Host for Ticks and Tick-Borne Pathogens in Peri-Urban Environments, Hungary
Abstract
European badgers are opportunistic animals that could serve as important hosts in the life cycle of hard ticks (Acari: Ixodidae) when entering peri-urban and urban environments. In this study, ticks and spleen samples were collected from badgers (Meles meles) found as roadkill between 2020 and 2021 in peri-urban habitats in Central Europe, Hungary. Altogether, 117 ticks, representing seven species (Ixodes ricinus, Ixodes kaiseri, Ixodes canisuga, Ixodes hexagonus, Haemaphysalis concinna, Alloceraea inermis and Dermacentor reticulatus) were removed from 49 badgers. Following assessment of suitability for obtaining spleen samples from the carcasses, DNA was extracted, and conventional or real-time PCRs were used to detect tick-borne pathogens in spleen samples of 38 badgers. Among protozoan parasites, two Babesia species, representing two phylogenetic groups, and Hepatozoon martis were identified. In addition, Candidatus Neoehrlichia lotoris, a novel Ehrlichia species (provisionally named as Candidatus Ehrlichia transdanubiensis), and Anaplasma phagocytophilum were detected in these tissue samples. The presence of these tick-borne pathogens in peri-urban mustelids indicate that they may provide a source for the infection of ixodid ticks which can in turn transmit these pathogens to humans or pet dogs in urban habitats. Thus, badgers pose an important epidemiological risk factor at the interface of sylvatic and synanthropic environments.
Summary
- •
European badgers prefer forested habitats, frequently close to urban areas. Middle-sized mammals, like badgers, are important hosts for all developmental stages of hard tick species as blood meal sources.
- •
We collected badger roadkills to broaden our knowledge on the epidemiological role of these mammals, focusing on the species of ticks and tick-borne pathogens for which they may serve as primary or accidental hosts.
- •
We found seven tick species and ticks of all developmental stages on badgers. Among these, two tick species are reported here for the first time from this host.
- •
With molecular methods (conventional and real-time PCRs), we detected six tick-borne pathogens in the spleen samples of badgers. Based on molecular and phylogenetical evidence, we identified a not-yet-named Ehrlichia species among the pathogens, tentatively called here as Candidatus Ehrlichia transdanubiensis.
- •
Our study provides new insights into the role of European badgers as suitable hosts for ticks and tick-borne pathogens near urban habitats.
1. Introduction
The European badger (Meles meles) is a middle-sized omnivorous member of the family Mustelidae (Mammalia: Carnivora). It is widely distributed in Europe and the western part of Asia [1], where in natural ecosystems, it has a preference for forested habitats [2, 3]. Being an opportunistic species, the European badger can adapt to and exploit urban ecosystems due to its omnivorous behaviour [2] and its ability to utilize anthropogenic waste [4]. Consequently, this mustelid species can overbridge natural and peri-urban or urban habitats [2].
Moreover, during the past few years, a tendency of increasing urbanization of badgers has been observed in several European countries. In fact, long after the establishment of urban fox populations in continental Europe, there is evidence suggesting that badgers may be following a similar trend and started their range expansion into cities through the colonization of urban habitats [5, 6]. The European badger was a protected species in Hungary until 2001. Badger population increased by 60% between 1987 and 2000, and their area of occurrence has also expanded with occupying new habitats in the Hungarian Great Plain (southern Hungary) [7]. According to the Hungarian National Game Management Database, the estimated mean population size was ca. 35,500 individuals (2020–2021) [8].
European badgers can harbour a wide range of pathogens from viruses to bacteria to ectoparasitic arthropods [9]. Among ixodid ticks, all species reported so far from badgers in Europe, for instance, Ixodes ricinus, Ixodes canisuga, Ixodes kaiseri, Ixodes hexagonus, Haemaphysalis concinna and Rhipicephalus sanguineus s.l., can infest either carnivorous pet animals (dogs and cats) or humans [10, 11]. Tick-borne protozoan parasites (piroplasms) and bacteria (i.e. Neoehrlichia and Ehrlichia sp.) were also reported to be shared between badgers and dogs in Central Europe [12, 13]. In addition, zoonotic bacteria were shown to be present in badgers in Southern, Western and Northern Europe, including Anaplasma phagocytophilum and Borrelia spp. [14–16]. These data highlight the importance of studying badgers as carriers of pathogens that may represent health risk to humans or domestic animals, particularly in urban and peri-urban environments.
Therefore, the aim of this study was to investigate the role of European badgers living in peri-urban areas as hosts of ticks and tick-borne pathogens. Considering pathogens, badger tissue DNA extracts were screened for the presence of protozoan, apicomplexan parasites (Babesia and Hepatozoon spp.) and bacteria of the family Anaplasmataceae, which have high veterinary–medical importance in both domestic and wild-living carnivores and/or humans in Europe.
2. Materials and Methods
2.1. Sample Collection and Blood Smear Preparation
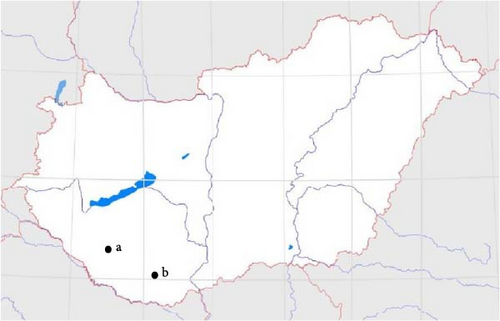
Road-killed badgers (n = 49) were collected in peri-urban areas in southwestern Hungary (Somogy [n = 26] and Baranya counties [n = 23], Figure 1) between 2020 and 2021 [17]. The fur covering of each animal was carefully checked for the presence of ticks with fine tweezers for ectoparasites and stored in 70% ethanol until identification [18, 19, 20]. In addition, when the state of the carcasses allowed (sometimes the collisions highly damaged the spleen), peppercorn-sized spleen samples were collected in microcentrifuge tubes from 38 badgers with NaClO-disinfected forceps and scissors. When it was possible, blood was also collected from the heart in EDTA tubes for morphological screening of intracellular forms of bacteria and protozoon. Blood samples were kept in the fridge (4°C) until blood smears were made from the nonhaemolyzed samples (n = 21) and thereafter stored at −20°C for further use as were the spleen samples. Blood smears were prepared, fixed with ethanol, stained with Giemsa and examined by a Leica DM 2000 light microscope on 400× magnification (Leica Microsystems, Wetzlar, Germany) [21].
2.2. DNA Extraction
DNA was extracted from the spleen samples by a QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany) following the manufacturers’ protocol with an overnight lysis. DNA samples were frozen (−20°C) until further use.
2.3. Molecular Investigation and Sequencing
Oligonucleotides and conditions of PCRs are summarized in Table 1, and reaction components are listed in Supporting Information 1: Appendix S1. During these tests, sequence-verified DNA of Babesia, Anaplasma and Hepatozoon spp. served as positive control. PCR products were sequenced by BIOMI Ltd. (Gödöllő, Hungary). Sequences were manually checked with BioEdit [28] and then compared to GenBank sequences by nucleotide BLASTn program (https://blast.ncbi.nlm.nih.gov). Sequences from this study, representing each genotype and species, were submitted to GenBank.
| Target | Product size (bp) | Primer name | Primer sequence | Thermal profile | Reference |
|---|---|---|---|---|---|
| Piroplasm 18S rRNA | ~500 | BJ1 | 5′-GTC TTG TAA TTG GAA TGA TGG-3′ | 95°C for 10 min; 40× (95°C for 30 s, 54°C for 30 s, 72°C for 40 s); 72°C for 5 min | [22] |
| BN2 | 5′-TAG TTT ATG GTT AGG ACT ACG-3′ | ||||
| Hepatozoon 18S rRNA | ~650 | HepF | 5′-ATA CAT GAG CAA AAT CTC AAC-3′ | 95°C for 5 min; 35× (95°C for 40 s; 57°C for 40 s; 72°C for 60 s); 72°C for 7 min | [23] |
| HepR | 5′-CTT ATT ATT CCA TGC TGC AG-3′ | ||||
| Anaplasmataceae 16S rRNA | ~350 | EHR-16sD | 5′-GGT ACC YAC AGA AGA AGT CC-3′ | 95°C for 10 min; 40× (95°C for 30 s, 55°C for 30 s, 72°C for 45 s); 72°C for 5 min | [24] |
| EHR-16sR | 5′-TAG CAC TCA TCG TTT ACA GC-3′ | ||||
| Anaplasma phagocytophilum GroEL | ~570 | EphplGroEL (569)F | 5′-ATG GTA TGC AGT TTG ATC GC-3′ | 95°C for 5 min; 40× (95°C for 30 s, 56°C for 40 s, 72°C for 1 min); 72°C for 7 min | [25] |
| EphGroEL (1142)R | 5′-TTG AGT ACA GCA ACA CCA CCG GAA-3′ | ||||
|
~1350 | Ehr1 | 5′-GAA CGA ACG CTG GCG GCA AGC-3′ | 95°C for 5 min; 40× (95°C for 30 s, 63°C for 40 s, 72°C for 1 min); 72°C for 7 min | [26] |
| Ehr6 | 5′-GAC CCA ACC TTA AAT GGC TGC-3′ | ||||
| ~1350 | Ehr7 | 5′-TAA CAC ATG CAA GTC GAA CG-3′ | 95°C for 5 min; 40× (95°C for 30 s, 60°C for 40 s, 72°C for 1 min); 72°C for 7 min | ||
| Ehr8 | 5′-CTT CGA GTT AAG CCA ATT CC-3′ | ||||
|
~1350 | HS1-f | 5′-CGY CAG TGG GCT GGT AAT GAA-3′ | 95°C for 5 min; 40× (95°C for 30 s, 55°C for 40 s, 72°C for 1 min); 72°C for 7 min | |
| HS6-r | 5′-CCW CCW GGT ACW ACA CCT TC-3′ | ||||
| ~1350 | HS3-f | 5′-ATA GTY ATG AAG GAG AGT GAT-3′ | 95°C for 5 min; 40× (95°C for 30 s, 50°C for 40 s, 72°C for 1 min); 72°C for 7 min | ||
| HSVR | 5′-TCA ACA GCA GCT CTA GTW G-3′ | ||||
| Anaplasma phagocytophilum msp2 qPCR | ~77 | ApMsp2f | 5′-ATG GAA GGT AGT GTT GGT TAT GGT ATT-3′ | 95°C for 20 s; 40× (95°C for 3 s, 60°C for 30 s) | [27] |
| ApMsp2r | 5′-TTG GTC TTG AAG CGC TCG TA-3′ | ||||
| ApMSP2p probe | 5′-Fam-TGG TGC CAG GGT TGA GCT TGA GAT TG-Tamra-3′ | ||||
2.4. Phylogenetic Analysis
All sequences retrieved from the GenBank and included in the phylogenetic analyses had 98%–100% coverage (i.e. aligned with a near-identical length and starting position) with sequences from this study. This dataset was resampled 1000 times to generate bootstrap values. Phylogenetic analysis was conducted by using the maximum likelihood method and Jukes–Cantor (Babesia 18S rRNA gene), Tamura–Nei (Anaplasma 16S rRNA gene) and Kimura 2 (Anaplasma GroEL gene) models according to the best fit selection with the program MEGA 7.0 [29].
3. Results
3.1. Tick Diversity
Altogether, 117 ticks were removed from the badgers, belonging to seven species (I. ricinus, I. kaiseri, I. canisuga, I. hexagonus, H. concinna, Alloceraea inermis and Dermacentor reticulatus). The most prevalent tick species was I. ricinus (n = 35) followed by I. kaiseri (n = 32), H. concinna (n = 20), I. canisuga (n = 15), I. hexagonus (n = 8), D. reticulatus (n = 5) and A. inermis (n = 4) (Table 2). The majority of these species were represented by nymphs (n = 56) and females (n = 48), but larvae (n = 4) and males (n = 9) also infested badgers in the case of three tick species (Table 2). The maximum intensity (maximum tick number/host) of tick infestation was seven individuals per host (sample ID: B5) but most badgers (n = 18) only harboured a single tick individual (mean intensity) (Supporting Information 2: Table S2).
| Species | Larva | Nymph | Female | Male | Sum |
|---|---|---|---|---|---|
| Ixodes ricinus | 1 | 8 | 23 | 2 | 35 (30%) |
| Ixodes kaiseri | 1 | 21 | 10 | — | 32 (27%) |
| Ixodes canisuga | — | 6 | 11 | — | 15 (13%) |
| Ixodes hexagonus | — | 6 | 2 | — | 8 (7%) |
| Haemaphysalis concinna | 2 | 14 | 2 | — | 20 (17%) |
| Alloceraea inermis | — | 1 | — | 3 | 4 (3%) |
| Dermacentor reticulatus | — | — | — | 4 | 5 (4%) |
| Sum | 4 | 56 | 48 | 9 | 117 (100%) |
- Note: The bold values seperate the summarized numbers from the normal individual numbers.
3.2. Tick-Borne Pathogens in Badger Blood and Spleen Samples
None of the blood smears evaluated in this study showed the presence of protozoan parasites or morulae of bacterial origin in red or white blood cells. However, molecular detection of Babesia, Hepatozoon, Anaplasma and Ehrlichia spp. (Table 3) was successful.
| Piroplasm | Hepatozoon sp. | Anaplasmataceae | Anaplasma phagocytophilum msp2 qPCR | Anaplasma phagocytophilum GroEL | Anaplasma phagocytophilum 16S nested | Anaplasma phagocytophilum GroEL nested |
|---|---|---|---|---|---|---|
| Positive/tested/prevalence % | ||||||
| 34/38/89.4% | 1/38/2.6% | 20/38/53% | 2/38/5.2% | 2/38/5.2% | 11/38/28.9% | 8/38/21% |
The prevalence of piroplasms in the spleen samples was 89.4% (34/38). Sequencing verified four different Babesia genotypes. Babesia sp. badger type A (OR144369) was found in 12 animals, and in 11 animals, three other genotypes of a different piroplasm were identified (OR144370, OR144372 and OR144373).
The spleen sample of one badger was positive for Hepatozoon sp., and sequencing identified Hepatozoon martis (OR144422).
In three samples Can. Neoehrlichia lotoris (OR158049), in 12 samples Ehrlichia sp. (OR158050), in one sample A. phagocytophilum (OR158052) and in another, a new Ehrlichia genotype (OR158051) was identified.
Samples of two badgers were positive for A. phagocytophilum with PCR detecting the GroEL gene of this species.
Eleven (27.5%) and eight (20%) badger samples were positive in two nested PCRs that target the 16S rRNA and GroEL genes, respectively, of A. phagocytophilum and closely related pathogens (Table 4).
| Target | Pathogen | Host | Samples | Number of genotypes (%) | Acc. number |
|---|---|---|---|---|---|
| Piroplasm 18S rRNA | Babesia sp. badger type A | Meles meles | BL1, BL5, BL9, BL10, BL21, BL24, BL28, BL35-39 | 12/50% | OR144369 |
| Babesia sp. new genotype 1 | Meles meles | BL4, BL18, BL22, BL26 | 4/16.5% | OR144370 | |
| Babesia sp. new genotype 1 | Martes foina | BL16 | 1/4% | OR144371 | |
| Babesia sp. new genotype 2 | Meles meles | BL12, BL25, BL33, BL34 | 4/16.5% | OR144372 | |
| Babesia sp. new genotype 3 | Meles meles | BL14, BL17, BL32 | 3/13% | OR144373 | |
| Hepatozoon 18S rRNA | Hepatozoon martis isolate | Meles meles | BL6 | 1/100% | OR144422 |
| Anaplasmataceae 16S rRNA | Candidatus Neoehrlichia lotoris | Meles meles | BL7, BL39, BL27 | 3/17.6% | OR158049 |
| Ehrlichia sp. | Meles meles | BL11, BL15, BL17, BL22, BL23, BL24, BL28, BL29, BL32, BL35, BL37, BL38 | 12/70.6% | OR158050 | |
| Ehrlichia sp. new genotype | Meles meles | BL40 | 1/5.9% | OR158051 | |
| Anaplasma phagocytophilum | Meles meles | BL19 | 1/5.9% | OR158052 | |
| Anaplasma phagocytophilum GroEL | Anaplasma phagocytophilum ecotype I | Meles meles | BL9 | 1/100% | OR167111 |
| Anaplasma phagocytophilum GroEL (nested) | Can. Ehrlichia transdanubiensis | Meles meles | BL7 | 1/20% | OR597008 |
| Can. Ehrlichia transdanubiensis | Meles meles | BL30 | 1/20% | OR597009 | |
| Can. Ehrlichia transdanubiensis | Meles meles | BL35, BL36 | 2/40% | OR597010 | |
| Can. Ehrlichia transdanubiensis | Meles meles | BL38 | 1/20% | OR597011 | |
| Anaplasma phagocytophilum 16S (nested) | Can. Ehrlichia transdanubiensis | Meles meles | BL7 | 1/10% | OR567853 |
| Can. Ehrlichia transdanubiensis | Meles meles | BL17, BL24 | 2/20% | OR567854 | |
| Can. Ehrlichia transdanubiensis | Meles meles | BL23, BL30, BL35, BL36, BL37 | 5/50% | OR567856 | |
| Can. Ehrlichia transdanubiensis | Meles meles | BL40 | 1/10% | OR567857 | |
| Anaplasma phagocytophilum | Meles meles | BL19 | 1/10% | OR567855 | |
- Note: An additional stone marten (Martes foina) spleen sample is also presented here because it was used in the phylogenetic tree (Figure 1).
4. Discussion
The number of studies published on ixodid ticks and tick-borne pathogens from European badgers is limited [10, 11, 16, 30–32]. In this study, all hard tick species reported so far from this host were found, except R. sanguineus. On the other hand, this is the first report of A. inermis and D. reticulatus on European badgers. These collected tick species can serve as vectors of human and companion animal-related pathogens, such as I. ricinus and I. hexagonus for Borrelia burgdorferi s.l. and D. reticulatus for Babesia canis.
The most prevalent genotype of piroplasm was Babesia sp. badger type A (OR144369), which was present in 12 animals. This piroplasm belongs to the Babesia microti group, as reflected by the phylogenetic analysis (Figure 2). This variant was also reported, among others, from badgers in Spain (KT223484), the United Kingdom (KX528553) and Hungary (KX218234), as well as from dogs in Hungary (MG778913) [11, 33, 34].
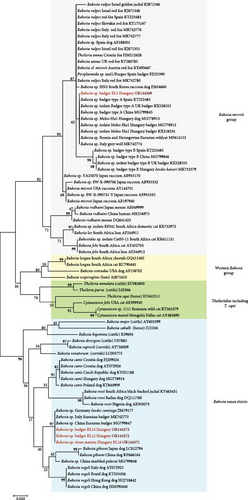
In further 11 animals, three genotypes of a different piroplasm were identified (OR144370, OR144372 and OR144373). These clustered in the phylogenetic group of Babesia sensu stricto (Figure 2). In Europe, to the best of our knowledge, this not-yet-named Babesia species has only been reported from badgers in Italy (MK742775) and from I. canisuga ticks in Germany (JX679177) [35, 36]. These findings on piroplasms confirm that at least two different unnamed Babesia species circulate in badgers and their ticks.
We found Hepatozoon martis (OR144422) in one European badger spleen sample. Hepatozoon martis has recently been reported in stone martens (Martes foina) and beech martens (Martes martes) from Hungary (OM256567 and OL960184) [37]. Thus, the present finding attests that badgers are also susceptible for H. martis.
In the general PCR for Anaplasmataceae pathogens, the rate of positivity was 53% (20 out of 38 badgers). Can. Neoehrlichia lotoris (OR158049) was identified in three samples, Ehrlichia sp. (OR158050) in 12 samples, A. phagocytophilum (OR158052) in one sample and a new Ehrlichia genotype (OR158051) in one sample. Interestingly, this new Ehrlichia genotype had a high (up to 100%) rate of identity to Ehrlichia spp. from different hosts, that is, a Hungarian dog (MH020203) [12], a badger sampled in Spain (KR262717) [38] and an Australian western quoll (Dasyurus geoffroii, MW633161) [39].
Samples from two badgers were positive for A. phagocytophilum. The GroEL sequence of this isolate (OR167111) indicated that it belongs to the first, most pathogenic and zoonotic ecotype and has 100% identity to A. phagocytophilum detected in different hosts, for example, a dog in Russia (MH535862) or Hungarian urban hedgehogs and associated I. ricinus ticks (MF372791 and MF372779) [40].
Anaplasma phagocytophilum (OR567855) and four genotypes of Ehrlichia species (OR567853, OR567854, OR567856 and OR567857) were shown to be present in these samples with the 16S rRNA nested reactions. Similarly, in the GroEL PCR, four new Ehrlichia sp. genotypes (OR597008, OR597009, OR597010 and OR597011) were demonstrated (Table 4).
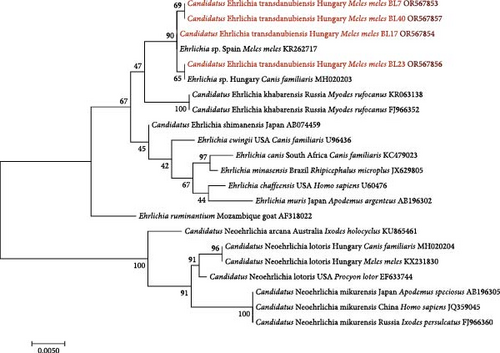
Ehrlichia sp. 16S rRNA sequences had high (99.76%–100%) rate of identity to two other Ehrlichia sp. sequences in GenBank: one from a Hungarian dog sampled at a nearby location in southwestern Hungary (MH020203) and the other collected from a European badger in Spain (KR262717). Based on both molecular and phylogenetic analyses, the most closely related species to this badger isolate is Can. Ehrlichia khabarensis, with 98.76% sequence identity. The 16S rRNA sequence of A. phagocytophilum had the highest (99.92%) identity to sequences from different vectors (ticks), hosts (e.g. dogs) and locations (e.g. South Africa and China) (MK814402-06 and OM569668) [41, 42]
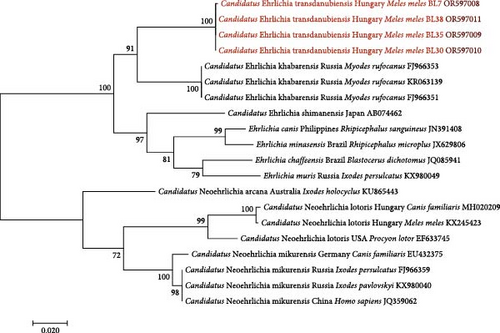
The four Ehrlichia sp. GroEL sequences showed the highest (92.42%) identity to three Can. Ehrlichia khabarensis sequences from the Russian Far East (FJ966351, FJ966353 and KR063139) [26, 43] The latter species was reported to have morulae in the spleen and liver cells of rodents, and its life cycle (including the vector) remains to be clarified [43].
The Ehrlichia sp. 16S rRNA phylogenetic tree of these genotypes from badgers reflected that they belong to the same clade (Figure 3) as Spanish and Hungarian samples (KR262717 and MH020203) [12, 44], and with low (47%) bootstrap support, they clustered separately from Can. Ehrlichia khabarensis sequences (KR063138 and FJ966352) [26, 43]. The phylogenetic tree based on the GroEL gene confirmed this separate clustering with high (88%) support between the badger-associated new Ehrlichia sp. and the clade of Can. Ehrlichia khabarensis (FJ966351, FJ966353 and KR063139) [26, 43] (Figure 4). Thus, based on sequence comparisons and phylogenetic relationships, the Ehrlichia sp. identified in badgers during the present study most likely represents a new species and was provisionally named as Can. Ehrlichia transdanubiensis, according to the Transdanubian region where most samples containing its genotypes originated in this study, as well as previous findings (8).
5. Conclusion
We found seven ixodid tick species (I. ricinus, I. kaiseri, I. canisuga, I. hexagonus, H. concinna, A. inermis and D. reticulatus) on European badgers. This study added two new hard tick species to those associated with European badgers. These were A. inermis and D. reticulatus. The range of tick-borne pathogens for which peri-urban badgers are hosts was also elucidated here, including two Babesia spp., Hepatozoon martis, Can. Neoehrlichia lotoris, A. phagocytophilum (zoonotic ecotype) and a novel Ehrlichia species (Can. Ehrlichia transdanubiensis). Among these pathogens, new genotypes broadened the spectrum of hitherto known variants of pathogens compare to other studies. These findings highlight the importance of monitoring peri-urban and urban badgers as hosts of ticks and tick-borne pathogens, thus raising awareness in this context. Further monitoring will help to reduce epidemiological risks posed by their presence in human settlements and by their consequent indirect contact with humans and companion carnivores via ticks.
Ethics Statement
All animals from which samples were collected during this study were found as roadkill. The utilization of cadavers for scientific purposes was in accordance with the Hungarian government decree 71/2015. (III.30.). No vertebrate animals were caught or restrained for sample collection; therefore, no ethical permission was needed.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding
This study was funded by the Hungarian Research Network (HUN-REN) (Project No. 1500107). This study served as an initiative for project STARTING-150266 provided by the Ministry of Culture and Innovation of Hungary from the National Research, Development and Innovation Fund.
Acknowledgments
The authors are grateful to Ciara Reynolds, DVM, for language editing. The authors are grateful to Gábor Majoros, DVM, PhD, for his indispensable contribution in the blood smear evaluation.
Supporting Information
Additional supporting information can be found online in the Supporting Information section.
Open Research
Data Availability Statement
The sequences in the study were uploaded to GenBank. The PCR reactions were from formerly published resources (cited in the study).




