Genome-Wide Mapping of Bivalent Histone Modifications in Hepatic Stem/Progenitor Cells
Abstract
The “bivalent domain,” a distinctive histone modification signature, is characterized by repressive trimethylation of histone H3 at lysine 27 (H3K27me3) and active trimethylation of histone H3 at lysine 4 (H3K4me3) marks. Maintenance and dynamic resolution of these histone marks play important roles in regulating differentiation processes in various stem cell systems. However, little is known regarding their roles in hepatic stem/progenitor cells. In the present study, we conducted the chromatin immunoprecipitation (ChIP) assay followed by high-throughput DNA sequencing (ChIP-seq) analyses in purified delta-like 1 protein (Dlk+) hepatic stem/progenitor cells and successfully identified 562 genes exhibiting bivalent domains within 2 kb of the transcription start site. Gene ontology analysis revealed that these genes were enriched in developmental functions and differentiation processes. Microarray analyses indicated that many of these genes exhibited derepression after differentiation toward hepatocyte and cholangiocyte lineages. Among these, 72 genes, including Cdkn2a and Sox4, were significantly upregulated after differentiation toward hepatocyte or cholangiocyte lineages. Knockdown of Sox4 in Dlk+ cells suppressed colony propagation and resulted in increased numbers of albumin+/cytokeratin 7+ progenitor cells in colonies. These findings implicate that derepression of Sox4 expression is required to induce normal differentiation processes. In conclusion, combined ChIP-seq and microarray analyses successfully identified bivalent genes. Functional analyses of these genes will help elucidate the epigenetic machinery underlying the terminal differentiation of hepatic stem/progenitor cells.
1. Introduction
Before 2000, most research on liver development and differentiation was performed using morphological approaches in knockout mice [1]. Thus, many transcription factors with important roles in hepatocyte and cholangiocyte differentiation have been reported [2, 3]. Advances in cell sorting technology since the beginning of the 21st century have led to progress in the isolation and identification of hepatic stem/progenitor cells [4], and it has become possible to analyze signal transduction pathways and molecules involved in the maintenance and/or differentiation of stem cells [5–7].
Epigenetic mechanisms, including DNA methylation and histone modification, are essential for cell fate decisions and differentiation during embryogenesis [8]. Particularly, histone modifications are dynamically regulated by enzymes that add or remove these modifications [9], and these modifications have been described as extremely important in developmental processes in both the liver and pancreas [10]. Although polycomb group (PcG) proteins are responsible for transcription-repressive histone H3 trimethylation at lysine 27 (H3K27me3), trithorax group complexes (TrxG) are associated with transcription-active histone H3 trimethylation at lysine 4 (H3K4me3) [11]. In embryonic stem (ES) and tissue-specific stem cells, the promoter regions of genes that regulate differentiation contain “bivalent domains” with both the H3K27me3 and H3K4me3 [12]. This configuration is believed to allow cell fate determination and differentiation to rapidly begin in any direction in response to intracellular and extracellular signals.
Here, we aimed to elucidate the mechanisms through which histone modifications regulate differentiation from the viewpoint of bivalent domains in normal hepatic stem/progenitor cells. We performed the chromatin immunoprecipitation (ChIP) assay followed by high-throughput DNA sequencing (ChIP-seq) analyses in delta-like 1 protein (Dlk+) hepatic stem/progenitor cells using anti-H3K4me3 and anti-H3K27me3 antibodies. Next, we performed microarray analyses using RNA isolated from Dlk+ and differentiated cells. A comprehensive analysis of these data allowed for determination of bivalent genes and elucidation of epigenetic regulatory machinery of the differentiation process in hepatic stem/progenitor cells.
2. Materials and Methods
2.1. Mice
Pregnant C57BL/6 mice were purchased from Japan SLC (Hamamatsu, Japan). They were bred and maintained in accordance with our institutional guidelines for the use of laboratory animals.
2.2. Purification and Culture of Dlk+ Cells
Dlk+ cells were prepared from embryonic day (ED) 14.5 fetal livers, as described previously [13]. Briefly, cells were stained with an anti-Dlk antibody (MBL, Nagoya, Japan) followed by exposure to anti-rat IgG-conjugated magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany). Dlk+ cells were corrected by passing them through cell separation columns under a magnetic field (Miltenyi Biotec).
2.3. Colony Assays and Terminal Differentiation Experiments
Dlk+ cells (1 × 103 cells/well) were plated on collagen type IV-coated 6-well plates (Becton Dickinson, Franklin Lakes, NJ) and cultured as described elsewhere [13]. At least three independent experiments were performed for colony assays. To evaluate the ability to differentiate into hepatocytes, Dlk+ cells were placed on an Engelbreth-Holm-Swarm (EHS) gel (Becton Dickinson) in the presence of oncostatin M (OSM, R&D Systems, Minneapolis, MN). Similarly, the cells were placed on collagen type I gel (Nitta Gelatin, Osaka, Japan) in the presence of tumor necrosis factor- (TNF-) α (PeproTech, Rocky Hill, NJ) to examine their potential to differentiate into cholangiocytes.
2.4. Flow Cytometry
To examine the purity of Dlk+ cells sorted by magnetic cell separation, the cells were stained with a phycoerythrin-conjugated anti-Dlk antibody (MBL) for 30 min on ice. Labeled cells were resuspended in PBS with 1% fetal bovine serum. Propidium iodide (1 μg/ml) was added for the elimination of dead cells. Cell analysis and sorting were performed using FACSCanto and FACSAria II (BD Biosciences, San Jose, CA).
2.5. Immunocytochemistry
Colonies derived from Dlk+ cells were stained with rabbit anti-albumin (Alb) (GeneTex, San Antonio, TX) and mouse anti-cytokeratin 7 (CK7) (DakoCytomation, Fort Collins, CO) antibodies followed by Alexa Fluor 555-conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR) and Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes) antibodies, respectively. The absolute number of Alb+CK7+ bipotent cells in each large colony at day 5 of culture was determined in at least 10 large colonies containing more than 100 cells. To examine cellular apoptosis, cells were stained with an anti-caspase 3 (CASP3; Millipore, Billerica, MA) antibody, followed by incubation with Alexa 555-conjugated IgG (Molecular Probes).
2.6. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
RNA was extracted using the RNeasy Mini kit (Qiagen, Valencia, CA), following the manufacturer’s protocol. cDNA synthesis was performed using the ThermoScript RT-PCR system (Invitrogen, Frederick, MD) with an oligo-dT primer. Quantitative RT-PCR was performed with an ABI PRISM 7300 Sequence Detection System (Applied Biosystems, Foster City, CA) using the Universal ProbeLibrary System (Roche Diagnostics, Mannheim, Germany) following the manufacturer’s protocol. The primer sequences are listed in Supplementary Table S1.
2.7. Lentiviral Production and Transduction
Lentiviral vectors (CS-H1-shRNA-EF-1a-EGFP) expressing short hairpin RNA (shRNA) targeting murine SRY-related high-mobility group (HMG) box 4 (Sox4, target sequence: sh-Sox4-1, 5 ′-ACCAACAACGCGGAGAACACT-3 ′; sh-sox4-2, 5 ′-GCGACAAGATTCCGTTCATCA-3 ′) and luciferase (Luc) were constructed using Gateway LR Clonase systems (Invitrogen, Carlsbad, CA). Recombinant lentiviruses were produced as described previously [14]. Cells were transduced with a lentiviral vector in the presence of protamine sulfate (10 μg/ml; Sigma, St. Louis, MO).
2.8. Western Blotting
Sox4-knockdown cells were selected via cell sorting for enhanced green fluorescent protein (EGFP) expression. Cells were lysed in RIPA buffer (50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA (pH 8.0), 1% Triton X-100, 0.1% sodium deoxycholate, and 0.1% SDS) with protease inhibitor cocktail (Roche). Lysates were then sonicated, separated by SDS-PAGE, and transferred to a PVDF membrane. Subsequently, they were subjected to Western blotting using anti-Sox4 (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-tubulin (Oncogene Science, Cambridge, MA) antibodies.
2.9. ChIP
ChIP assay was performed as described previously [15]. Briefly, cross-linked chromatin was sonicated into 200–500-base-pair fragments. The immunoprecipitated and input DNA was treated using anti-H3K4me3 (Millipore, Bedford, MA, USA) and anti-H3K27me3 antibodies (Millipore). Normal mouse IgG was used as a negative control. The immunoprecipitated and input DNA was treated with RNase A (Sigma-Aldrich) and proteinase K (Roche) and purified using the QIAquick PCR purification kit (Qiagen). Quantitative PCR was conducted using the ABI Prism 7300 Thermal Cycler with SYBR Premix Ex Taq II (Takara Bio, Otsu, Japan). The primer sequences used are listed in Supplementary Table S2.
2.10. ChIP-seq
The reads per million (RPM) mapped read values of the region from 2 kb upstream to 2 kb downstream to the transcription start site (TSS) of the immunoprecipitated samples were divided by the RPM of the corresponding input. The RPM values of the sequenced reads were calculated every 2,000-base-pair bins with a shifting size of 200 base pairs using BEDTools. The RPM values of the sequenced read were calculated using BEDTools. The RPM values of the sequenced reads were calculated using BEDTools and visualized using the Integrative Genomics Viewer (http://www.broadinstitute.org/igv). The ChIP-seq data obtained in this study were deposited in the DNA Data Bank of Japan (DDBJ, accession number DRA006858).
2.11. Microarray Analysis
Microarray analysis was conducted using SurePrint G3 Mouse GE microarray (Agilent Technologies, Santa Clara, CA, USA). The microarray was labeled, hybridized, washed, and scanned as described by the manufacturer. The expression value (signal) for each probe set was calculated using GeneSpring GX 12.0 (Agilent). Data were normalized using GeneSpring normalization algorithms (Agilent). Only statistically significant gene expression levels (P < 0.05) were recorded as being “detected” above background levels, whereas genes with expression levels lower than this statistical threshold were considered “absent.” To identify differentially expressed genes involved in the differentiation of Dlk+ cells, genes exhibiting greater than 2.0-fold changes were selected. Moreover, gene ontology (GO) annotations were performed using the GeneSpring annotation tool (Agilent). The significance of each term was determined using Fisher’s exact test and the Bonferroni adjustment for multiple testing. The raw data are available at Gene Expression Omnibus (GEO, accession number: GSE 114833).
2.12. Statistical Analysis
Data are presented as mean ± SEM. Statistical differences were analyzed using the Mann–Whitney U test. The correlation between histone marks and expression levels was analyzed using Pearson’s correlation analysis. P values less than 0.05 were considered statistically significant.
3. Results
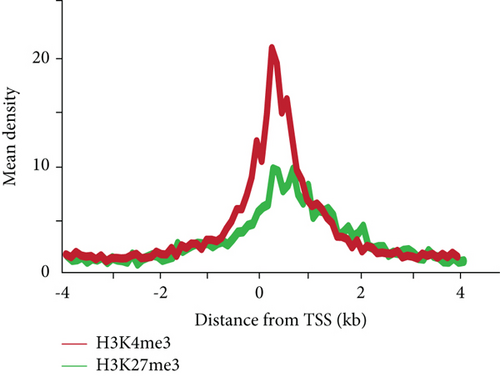
3.1. Genome-Wide Analyses of H3K4me3 and H3K27me3 in Hepatic Stem/Progenitor Cells
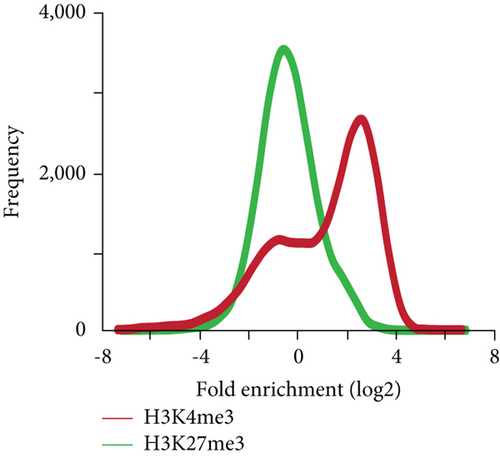
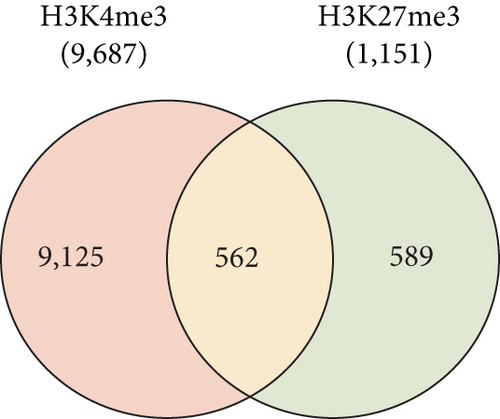
To gain an insight into the histone modification of hepatic stem/progenitor cells, we purified Dlk+ cells from fetal livers at ED 14.5 using magnetic-activated cell sorting (MACS). Flow cytometric analysis revealed that the purity of the sorted Dlk+ cells was approximately 90% (Supplementary Figure S1). To explore the genome-wide distribution of H3K4me3 and H3K27me3 in hepatic stem/progenitor cells, purified Dlk+ cells were subjected to ChIP-seq analysis. Although both the H3K4me3 and H3K27me3 peaks were observed in the slight downstream region of TSS, the H3K27me3 peaks had greater width and centrally depressed signals (Figure 1(a)). We focused on the region within 2.0 kb of the TSSs of the reference sequence genes (Figure 1(b)). Subsequently, ChIP-seq analysis successfully identified 9,687 genes with H3K4me3 enrichment and 1,151 genes with H3K27me3 enrichment greater than 2-fold (log2) of the input levels. In total, 562 genes possessed bivalent chromatin domains containing H3K4me3 and H3K27me3 marks (Figure 1(c)).
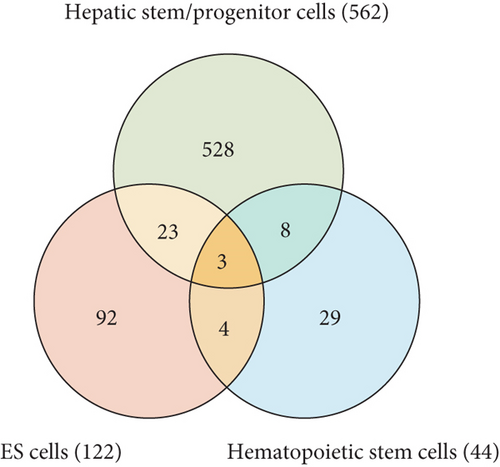
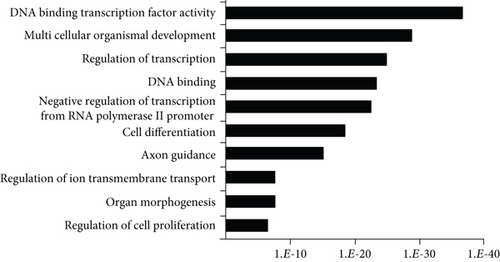
It has been reported that polycomb proteins Bmi1 and Ezh2 are required for self-renewal regulation in hepatic stem/progenitor cells as well as in pluripotent stem cells, such as ES cells, and hematopoietic stem cells [5, 13]. These findings suggest that common machinery plays an important role in differentiation regulation in ES cells and somatic stem cells. Therefore, we subsequently compared the list of these genes with the ChIP-seq data examined in mouse ES cells and human/mouse hematopoietic stem cells (HSCs) [16, 17]; 26 and 11 genes with bivalent promoters in hepatic stem/progenitor cells were overlapped with bivalent genes in ES cells and HSCs, respectively (Figure 1(d)). Among them, three genes, namely, Ebf1, Pax6, and HoxA1, were overlapped with genes with bivalent promoters in both ES cells and HSCs. To gain insights into their biological roles, we analyzed the 562 identified bivalent genes. Functional annotations based on GO revealed significant enrichment of genes belonging to the categories “DNA binding,” “transcription,” “development,” and “differentiation” (Figure 1(e)).
3.2. Comparison between Histone Marks and Gene Expression Levels
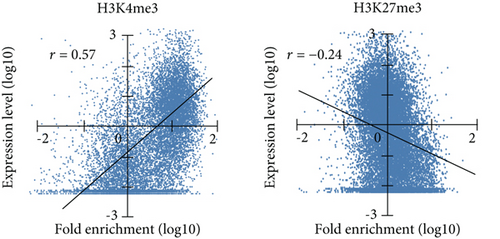
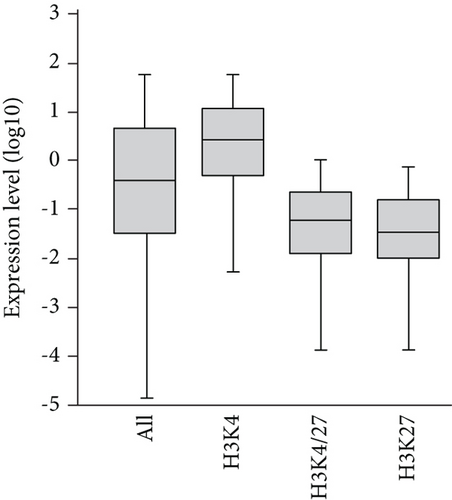
Initially, based on the microarray data of freshly purified Dlk+ cells, we examined the relationship between histone modification patterns and gene expression levels (Figure 2(a)). Pearson’s correlation coefficient analysis revealed a positive correlation between gene expression and H3K4me3 levels (r = 0.57) and a negative correlation between gene expression and H3K27me3 levels (r = −0.24). We compared expression levels among genes marked with H3K4me3, H3K27me3, and both H3K4me3 and H3K27me3 (bivalent) (Figure 2(b)). Genes with H3K4me3 marks showed higher expression levels, whereas those with H3K27me3 marks and bivalent marks (H3K4/27me3) displayed lower expression levels.
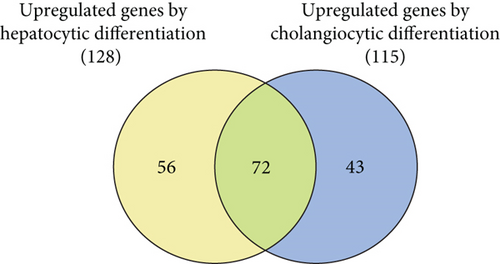
3.3. Identification of Bivalent Genes Showing Derepression during Differentiation
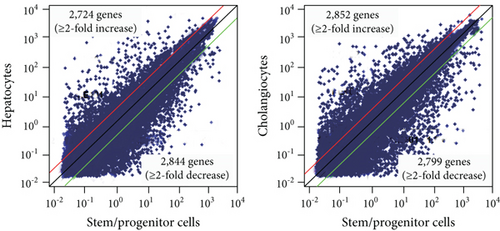
Next, we cultured Dlk+ cells on EHS gel in the presence of OSM to induce differentiation into mature hepatocytes and on collagen type I gel in the presence of TNF-α to selectively induce differentiation into cholangiocytes. mRNA extracted from these differentiated cells was subjected to microarray analysis. We successfully identified 2,724 upregulated and 2,844 downregulated genes after the differentiation of Dlk+ cells into hepatocytes (Figure 2(c)). Similarly, 2,852 genes were upregulated and 2,799 were downregulated after differentiation into cholangiocytes. Bivalent domains are considered to control the expression of developmental genes, thus allowing timely activation during the differentiation process. To gain insights into epigenetic differentiation regulators, we highlighted the bivalent genes that were derepressed following the differentiation of hepatic stem/progenitor cells. As a result, 128 and 115 genes were upregulated during differentiation into hepatocytes and cholangiocytes, respectively (Figure 2(d)). Among these, 72 genes were commonly upregulated during differentiation toward both the hepatocyte and cholangiocyte lineages (Supplementary Table S3).
3.4. Changes of Histone Marks in the Ink4a/Arf Gene Locus during Differentiation
Among the bivalent genes, the Ink4a/Arf locus was modified by bivalent histone marks in purified Dlk+ cells. Ink4a/Arf is an important target of PcG, and its transcriptional repression is essential for maintaining self-renewal in hepatic stem/progenitor cells [18]. Consistent with microarray data, real-time PCR analyses demonstrated that remarkable derepression of Ink4a/Arf was observed after the induction of hepatocytic and cholangiocytic differentiation (Supplementary Figure S2). ChIP analyses revealed high levels of both H3K4me3 and H3K27me3 marks at the Ink4a/Arf promoter in purified Dlk+ cells. However, H3K27me3 levels, but not H3K4me3 levels, were diminished at the locus in mature hepatocytes and cholangiocytes.
3.5. Bivalent Histone Modification on the Sox4 Gene Locus
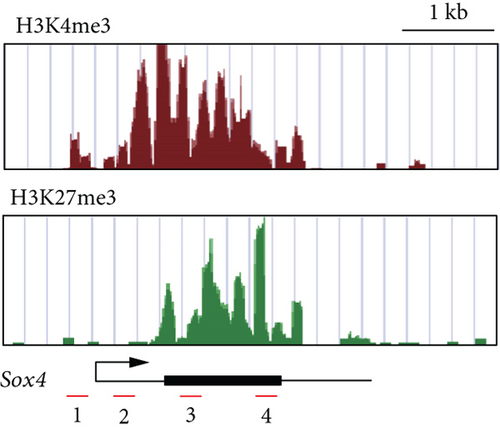
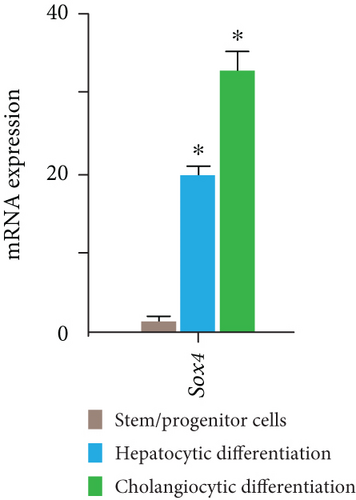
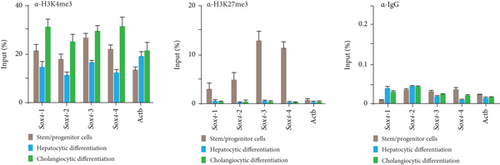
Similar to Ink4a/Arf, SRY-related HMG box 4 (Sox4) exhibited bivalent loci in purified Dlk+ cells (Figure 3(a)). Real-time PCR analyses demonstrated that Sox4 was significantly upregulated after differentiation toward the hepatocyte and cholangiocyte lineages (Figure 3(b)). Sox4 upregulation was more prominent in cholangiocytes than in hepatocytes. We next focused on the role of Sox4 in hepatic stem/progenitor cells. Sox4 upregulation was more prominent during cholangiocytic differentiation than during hepatocytic differentiation. Although high levels of both H3K4me3 and H3K27me3 were detected at the Sox4 promoter in purified Dlk+ cells, only H3K4me3 levels were increased at the Sox4 locus in terminally differentiated cells (Figure 3(c)). These findings indicate that the Sox4 expression level is tightly regulated through histone modifications in both stem/progenitor and terminally differentiated cells.
3.6. Loss-of-Function Assays of Sox4 in Hepatic Stem/Progenitor Cells
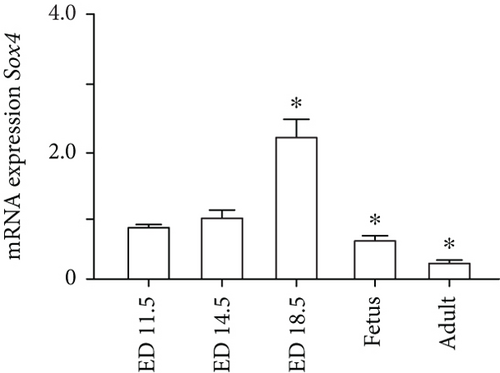
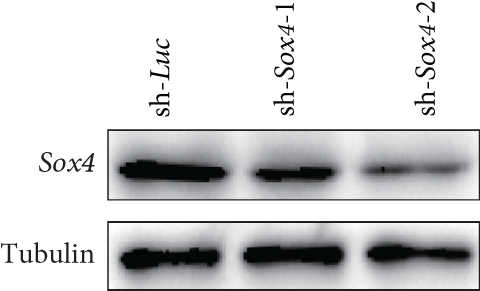
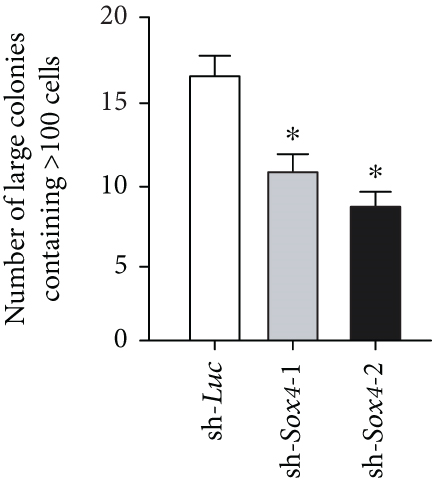
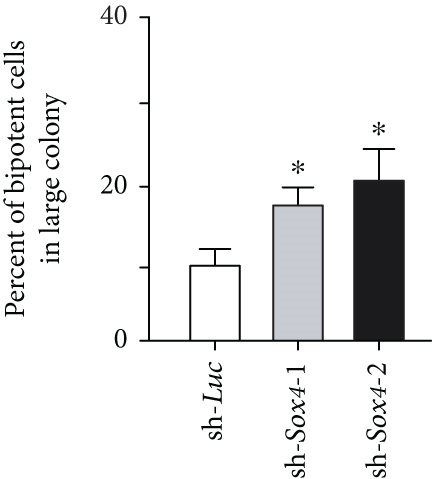
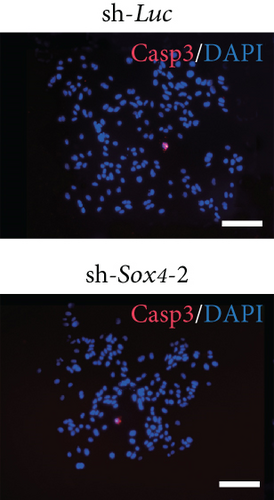
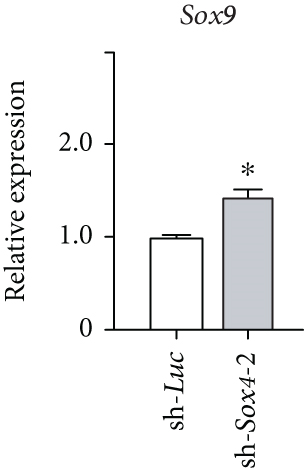
Quantitation of Sox4 mRNA extracted from fetal, neonatal, and adult livers using real-time RT-PCR demonstrated that the Sox4 expression achieved a peak in late-gestational fetal liver and decreased rapidly after that (Figure 4(a)). These findings indicated the possibility that derepression of Sox4 was essential for the normal differentiating process. These results prompted us to test the effects of Sox4 in a loss-of-function assay. Sox4 knockdown was confirmed using Western blotting (Figure 4(b)). Sox4 knockdown impaired colony formation and reduced both the number of large colonies containing >100 cells and the size of colonies (Figure 4(c)). Immunocytochemical analyses revealed an increase in the number of Alb+CK7+ bipotent cells in colonies derived from Sox4-depleted Dlk+ cells compared with the findings in control colonies on day 5 of culturing (Figures 4(d) and 4(e)). However, no remarkable differences were observed in a number of Casp3-positive cells in control and Sox4-knockdown colonies (Figure 4(f)). In addition, Sox4 knockdown resulted in a modest but significant increase in Sox9 mRNA expression (Figure 4(g)). Taken together, Sox4 is essential for the differentiation of Dlk+ cells into mature hepatocytes and cholangiocytes.
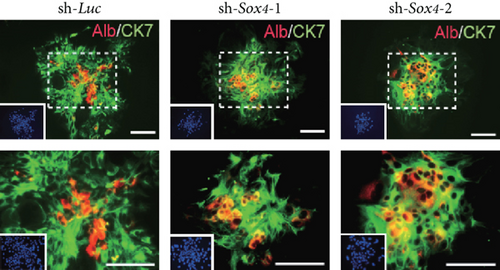
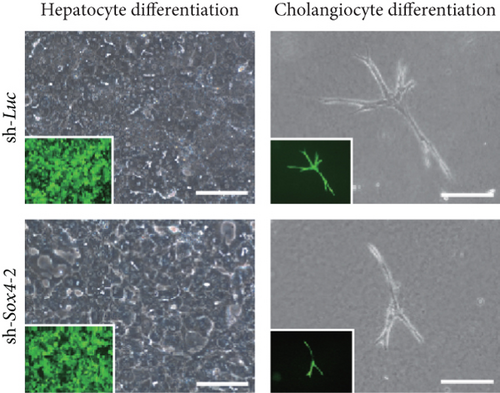
To selectively induce terminal hepatocyte maturation, we cultured Dlk+ cells in EHS gel supplemented with OSM. Multiple cell clusters with tight cell-cell contact were formed by day 4 of culturing. Compared with the findings for the control clusters, the Sox4-knockdown clusters lacked hepatocytes with highly condensed, granulated cytosol and clear round nuclei (Figure 4(h)). Concordant with these findings, real-time RT-PCR analyses revealed that hepatocyte-lineage markers, such as Alb, tyrosine aminotransferase (Tat), hepatocyte nuclear factor 3β (Hnf3β), and Hnf4α, were significantly downregulated in Sox4-depleted cells (Supplementary Figure S3(a)). In contrast, stem cell markers, such as alpha-fetoprotein (Afp), epithelial cell adhesion molecule (Epcam), and Bmi1, were significantly upregulated in Sox4-knockdown cells. Next, Dlk+ cells were cultured on collagen type I gel in the presence of TNF-α to selectively induce cholangiocytic differentiation. Sox4-depleted Dlk+ cells, but not control Dlk+ cells, failed to form tube-like structures (Figure 4(e)). As expected, real-time RT-PCR analyses demonstrated that cholangiocyte-lineage markers, such as CK7, integrin b4 (Itgb4), Hnf1β, and Hnf6, were significantly downregulated, and stem cell markers mentioned above were upregulated in Sox4-depleted cells (Supplementary Figure S3(b)). Taken together, Sox4 gene activation is essential for terminal differentiation toward the hepatocyte and cholangiocyte lineages.
4. Discussion
Bivalent domains simultaneously contain the opposing histone modification H3K4me3 and H3K27me3 and have been found in the promoters of a broad range of genes that regulate differentiation in ES cells [19]. This elegant mechanism maintains the cell’s undifferentiated state while permitting target gene expression to be rapidly activated or repressed when cell fate is determined by differentiation signals [20]. Bivalent domains are intimately involved in the balance between TrxG and PcG histone modifications. In fact, ES cells lacking components of the PcG complex reportedly lose the H3K27me3 histone modification, leading to derepression of bivalent genes [21, 22]. Hence, the undifferentiated state of ES cells is lost, and differentiation accelerates toward a certain cell lineage. Deficits in pluripotency and abnormal differentiation have also been reported in tissue-specific stem cells lacking Ezh2 [23, 24]. We previously reported that depleting Ezh2 in hepatic stem/progenitor cells eliminates their capacity of self-renewal and causes them to exhibit abnormal differentiation that is skewed toward the hepatocyte lineage [13]. This suggests that PcG plays an important role in repressing differentiation programs in stem/progenitor cells.
Recently, the popularization of the ChIP-seq analysis, which combines next-generation sequencers with ChIP, has facilitated comprehensive analyses to determine binding sites for transcription factors or enrichments for specific histone modifications [25]. However, few reports have comprehensively analyzed histone modifications in stem/progenitor cells. In the current study, we first performed ChIP-seq analyses using purified Dlk+ cells and antibodies against H3K4me3 and H3K27me3. After exploring conditions in which H3K4me3 levels best correlate with gene expression and H3K27me3 levels best inverse-correlate with gene expression, we decided to analyze genes with enrichment of >2-fold (log2) of input in the region within 2.0 kb of the TSSs. We subsequently identified 562 bivalent genes. Of these genes, 26 overlapped with bivalent genes in ES cells, 11 overlapped with bivalent genes in hematopoietic stem cells, and 3 were common to both. Differentiation of hepatic stem/progenitor cells appears to be controlled precisely through epigenetic regulations of bivalent genes. Although these genes are mainly specifically found in hepatic stem/progenitor cells, some of them are commonly found in different stem cell systems.
Next, we integrated ChIP-seq and microarray data to identify bivalent genes that are derepressed when differentiation is induced. Our results indicated that 72 genes were commonly upregulated during differentiation toward both the hepatocyte and cholangiocyte lineages. In fact, one of the genes was Ink4a/Arf, which is an important PcG target gene. We previously reported that derepression of the Ink4a/Arf locus in Bmi1−/− Dlk+ cells leads to impaired growth activity and self-renewal [18]. Conversely, Ink4a/Arf−/− Dlk+ cells exhibit remarkable stem/progenitor cell expansion. Further analyses are needed to examine whether 72 genes cited in this study are repressed to maintain pluripotency in hepatic stem/progenitor cells and transcriptionally activated via loss of repressive histone modifications for normal differentiation to proceed.
The 72 aforementioned genes include Sox4 and Sox11. Sox genes are transcription factors belonging to the HMG box superfamily. Some Sox genes are conserved across species, ranging from zebrafish to humans [26]. As their name suggests, Sox genes display homology to the DNA-binding HMG box domain of the sex-determination gene Sry (generally greater than 50% homology within the HMG box), and to date, more than 20 different Sox genes have been cloned from mice [27]. Sox4 plays an important role in regulating differentiation in neural stem and hematopoietic stem/progenitor cells; however, its function in hepatic stem/progenitor cells is unclear [28, 29]. Therefore, we performed a more detailed analysis of Sox4. Although Sox4 expression in Dlk+ cells was extremely low, levels of the transcriptionally repressive H3K27me3 histone modification decreased as the cells were induced to differentiate into hepatocytes and cholangiocytes, thereby leading to 20- and 33-fold increases in Sox4 expression, respectively. Lentiviral-knockdown of Sox4 in Dlk+ cells impaired terminal differentiation toward hepatocyte and cholangiocyte lineages and causes an increase in Alb+CK7+ progenitor cells in Dlk+ cell-derived colonies. Concordant with these findings, Poncy and colleagues reported that cholangiocyte differentiation and bile duct formation are perturbed in developing livers in Sox4-knockout mice [30]. In the present study, Sox4 knockdown resulted in modest but significant upregulation of Sox9 expression in Dlk+ cell-derived colonies. Given that Sox4 and Sox9 cooperatively regulate development of bile ducts [30], this change might be a compensatory increase. Further analyses are necessary to elucidate the role of Sox4 and interaction between Sox4 and Sox9 in the regulation of hepatic stem/progenitor cells.
Furuyama and colleagues reported that based on a genetic lineage tracing analysis in mice, hepatocytes and cholangiocytes differentiate from Sox9-negative hepatoblasts [31]. In the current study, the H3K27me3 modification at the Sox9 locus was slightly below the cutoff, and it was therefore not included as a bivalent gene. However, Sox9 expression in Dlk+ cells was extremely low, and we observed a greater than 5-fold increase in its expression after the induction of differentiation into hepatocytes or cholangiocytes. We previously revealed that Sox17 is a target of Bmi1 and that its overexpression in Dlk+ cells accelerates their differentiation [18]. These examples illustrate that Sox genes in Dlk+ cells are finely controlled at the transcriptional level via histone modifications and suggest that they play important roles in controlling the differentiation of hepatic stem/progenitor cells.
Finally, our findings demonstrated that several genes possess bivalent histone marks in hepatic stem/progenitor cells. Additionally, drastic changes in the expression of these genes caused by the loss of bivalent domains are closely associated with the normal differentiation process. Further analyses are necessary to determine the roles of the identified genes in hepatic stem cell systems.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work is partially supported by grants from the Japan Society for the Promotion of Science (JSPS, #16K09340) and the Program for Basic and Clinical Research on Hepatitis from Japan Agency for Medical Research and Development (AMED, #JP18fk0210014).
Open Research
Data Availability
The microarray and ChIP-seq data obtained in this study have been deposited in Gene Expression Omnibus (GEO, accession number: GSE 114833) and in the DNA Data Bank of Japan (DDBJ, accession number: DRA006858), respectively. Other data used to support the findings of this study are available from the corresponding author upon request.




