Is vertical transmission the only pathway for Rickettsia felis?
Abstract
The genus Rickettsia encompasses several species grouped into two main clusters, Typhus and the Transitional groups. The latter group contains Rickettsia felis, an endosymbiont of several arthropods with an uncertain human pathogenicity and whose most efficient transmission mechanism described thus far is transovarial. The aim of this study was to evaluate whether this pathway exists using phylogenetic analysis and partial sequences of the 17kDa and gltA genes and comparing them with host phylogeny using the cytb region. This is the first study that evaluates the vertical transmission of R. felis. In general, both phylogenies of R. felis showed no polytomies, as suspected if this pathway was the only pathway occurring. When phylogenies of the invertebrates and the gltA of R. felis were compared for strong coevolutionary insight, intricate relationships were observed, suggesting that other transmission pathways must occur, such as horizontal transmission. Further studies are needed to determine which other transmission routes occur in hematophagous arthropods.
1 INTRODUCTION
Flea-associated Rickettsiae represents one of the most neglected fields within rickettsiology (Blanton & Walker, 2017). Two groups are recognized: the Typhus group with a single species, Rickettsia typhi, the causal agent of endemic typhus, and the Transitional group, where Rickettsia asembonensis, Candidatus Rickettsia senegalensis (both nonpathogenic agents to date) and Rickettsia felis (species whose pathogenicity is under debate) are grouped (Blanton & Walker, 2017; Caravedo Martinez et al., 2021).
Rickettsia felis is the first agent described within the Transitional group in 1990 and has been isolated from the cat flea Ctenocephalides felis felis in Africa (Caravedo Martinez et al., 2021). This species represents a particular case because it has been detected using molecular methods in a wide range of arthropods, among which hematophagous groups, such as fleas and mosquitoes, stand out, as well as free-living groups, such as booklice (Psocodea) and moths (Lepidoptera) (Behar et al., 2010; Brown & Macaluso, 2016; de Araújo et al., 2021; Parola et al., 2016).
Originally identified as a flea endosymbiont, it was later incriminated as the causal agent of a febrile illness in patients from North America and Southeast Asia (Ye et al., 2021). Particularly, in Mexico, it was identified as a potential causative agent of pleuritis and hepatomegaly (Pérez-Osorio et al., 2008).
The debate regarding its pathogenicity begins with the exclusive identification of DNA of the agent in peripheral blood of individuals with febrile malaria-negative cases in Africa as well as in Southeast Asia (Keller et al., 2021; Labruna & Walker, 2014). However, efforts to isolate R. felis from blood samples have been unsuccessful. This is paradoxical given that the few isolated strains deposited in collections have been recovered from various species of fleas, spreading exclusively in arthropod cell lines (mosquitoes and ticks) (Luce-Fedrow et al., 2014).
During the last 10 years, experimental assays have been carried out, showing that transovarial transmission is an efficient mechanism for the proliferation of Rickettsia species (Behar et al., 2010; Driscoll et al., 2020). In this sense, it has been shown that R. felis can remain for up to 10 subsequent generations in flea colonies. This result has revealed that the possible subsistence mechanism of the agent is merely vertical in arthropod populations (Legendre & Macaluso, 2017; Reif & Macaluso, 2009). This has also been documented in other groups of tick endosymbionts, such as members of the genera Wolbachia, Francisella and Coxiella (Driscoll et al., 2020). Population genetics studies show a high degree of structuring of the populations of these microorganisms, with the presence of a high number of unique haplotypes that match the phylogeny of the invertebrates that host them (Gerth et al., 2013).
Considering that several species of arthropods have been recorded to be infected by R. felis, the aim of this study was to evaluate whether vertical transmission is the only transmission mechanism in R. felis, as has been previously considered. If this is true, strong coevolutionary signals should be observed among R. felis and their invertebrate host phylogenies. Additionally, no genetic structure should be present within each subgroup of invertebrates, observed as polytomies in the phylogeny.
2 MATERIALS AND METHODS
Available sequences of the 16S rDNA, 17kDa protein and gltA protein were obtained from GenBank; only sequences with geographic and host information were included (Supporting Information 1). Additionally, sequences for R. typhi were obtained and used as an outgroup. Each set of sequences was aligned by gene using the MUSCLE algorithm (Edgar, 2004) in AliView (Larsson, 2014). However, only alignments of the 17kDa and gltA proteins were used for further analysis since for these genes, there are sufficient sequences with the necessary information, such as geography and hosts (Supporting Information 2).
To corroborate clonal reproduction as a proxy to infer vertical transmission of R. felis, phylogenetic relationships were obtained by the Bayesian inference (BI) approach in MrBayes 3.2 (Ronquist et al., 2012) for each alignment since no concatenated alignments were possible due to no-match sequences per individual. The best-fit substitution model and partition scheme were calculated in PartitonFinder 2 with mrbayes models and the greedy scheme search (Guindon et al., 2010; Lanfear et al., 2017), considering the Bayesian information criterion. The phylogenetic hypothesis was inferred using the Markov chain Monte Carlo (MCMC) algorithm, with three hot chains and one cold chain in two independent runs of 10 million generations, sampling every 1000 iterations. The convergence of results was observed in Tracer 1.7 (Rambaut et al., 2018), considering ESS values above 200. The final topology was obtained by a majority rule consensus tree with a 25% burn-in.
If vertical transmission is the only pathway for R. felis, we hypothesize that the phylogeny of this endosymbiont matches the host phylogeny as a coevolution process. To assess this, we obtained available sequences of the cytb mitochondrial region of invertebrate hosts where sequences of R. felis were reported. In some cases, since the species of the arthropod was not available, the sequence of the nearest sister species was used according to the phylogenies of each group (Table 1). With these sequences, a new alignment, partition scheme, best-substitution model and BI phylogenetic approach were performed as previously described for R. felis. Additionally, to create a tanglegram and show how the phylogeny of the host relates to the phylogeny of R. felis, a new phylogenetic analysis was performed only with sequences of R. felis where the invertebrate host was specified.
| Recorded host | Host species used | Accession number |
|---|---|---|
| Aedes albopictus | MK241804 | |
| Anopheles punctipennis | Anopheles crucians | NC_057651 |
| Anopheles sinensis | MF322628 | |
| Archaeopsylla erinacei | LT703441 | |
| Ctenocephalides canis | LN899589 | |
| Ctenocephalides felis felis | NC_049858 | |
| Ctenophthalmus congeneroides | KM890737 | |
| Ctenophthalmus solutus | Ctenophthalmus agyrtes | KM890651 |
| Culex pipiens | KT851543 | |
| Ixodes granulatus | Ixodes scapularis | MZ645749 |
| Liposcelis bostrychophila | KT873486 | |
| Pulex echidnophagoides | Pulex irritans | LT797481 |
| Rhadinopsylla insolita | Rhadinopsylla dahurica | KM890715 |
| Rhipicephalus sanguineus s.l. | KF251029 | |
| Stenoponia sidimi | KM890611 | |
| Uranotaenia sapphirina | AF311256 | |
| Xenopsylla cheopis | MW310242 | |
| Xenopsylla ramesis | KM890637 |
3 RESULTS AND DISCUSSION
This communication represents the first evaluation of the vertical transmission of R. felis worldwide with in silico analyses. The 17 kDa alignment with 61 sequences of 206 base pairs was partition-analyzed as follows: Felsenstein's (1981) model (Felsenstein, 1981) for the first position, the Kimura (1980) model (Kimura, 1980) with invariant models for the second position and the Hasegawa et al. (1985) model (Hasegawa et al., 1985) for the third position (BIC = 1609.06). The gltA alignment with 109 sequences of 289 base pairs was also analyzed by codons as follows: Felsenstein's (1981) model for the first and third codons and the Hasegawa et al. (1985) model for the second ones (BIC = 2,669.298) (Supporting Information 3).
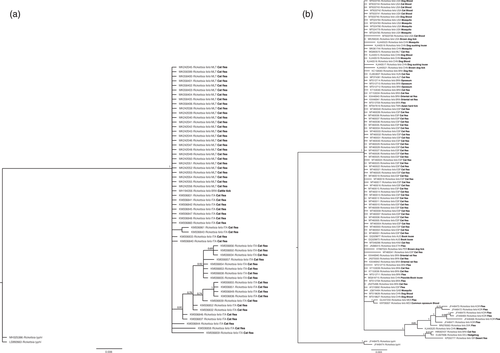
In general, similar patterns were obtained for both phylogenetic analyses and genetic distances. For both genes (17kDa and gltA), no polytomy was shown in the R. felis clade (Figure 1), as must be suspected when vertical transmission is the only pathway for spread (Martins et al., 2021). Instead, for the total of 289 bp of the gltA gene, 82% (237 bp) were conserved sites, 20% (57 bp) were variable sites, and 4% (12 bp) were single variable sites. For the 206 bp of the 17kDa gene, the values were similar, with 82% (170 bp) conserved, 17% (36 bp) variable and 2% (5 bp) single variable sites.
When the relationships between the gltA phylogeny and its hosts were compared, no coevolution signal was observed, even when a higher host phylogeny level was considered (Figure 2). The clearest evidence was found within the order Diptera, where all host species matched subgroup 1, except Culex pipiens, which matched subgroup 3. Another clear inconsistency is shown within the order Siphonaptera, where the host species matched all four subgroups, even within each species, such as Stenoponia sidimi and Rhadinopsylla cheopis, which matched subgroups 2 and 3, and C. felis, which matched subgroups 1 and 4 (Figure 2).
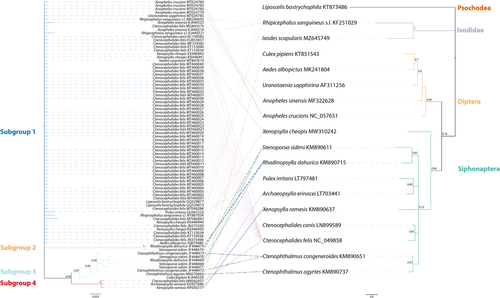
As previously reported for another bacterial endosymbiont of arthropods, those of the genus Wolbachia, if R. felis strains are only inherited by vertical transference, this should be traceable by mapping R. felis strains as traits onto the host phylogeny (Gerth et al., 2013; Martins et al., 2021). However, the lack of correspondence between host and R. felis phylogenies suggests not only a scattered random distribution among the hosts but also that other mechanisms are involved in the transmission of R. felis.
To explain the diversity and lack of coevolutionary signals between R. felis and its hosts, other evolutionary events, such as duplication, host switching or transfer, had to have occurred in both invertebrate and/or vertebrate hosts. One possible explanation is the possibility of horizontal transmission of R. felis, for example, through co-feeding, which would also explain its presence in other hematophagous arthropods with no confirmed vector role (Fecchio et al., 2018).
Finally, it is important to highlight that more studies must focus on determining alternative routes for infection by R. felis in hematophagous arthropods, with the aim of increasing the knowledge of this pathogenic-like Rickettsiae species and elucidating its transmission routes.
ACKNOWLEDGEMENT
This research was funded by Fondo para el Desarrollo del Conocimiento (FONDEC) No. FNV-2021-02 and Universidad Autónoma de Querétaro (UAQ).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required in this rapid communication since no animal samples were used.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the results of this study are available in GenBank. Accession numbers are available in all figures.




