Ecological super-spreaders drive host–range oscillations: Omicron and risk space for emerging infectious disease
Abstract
The unusual genetic diversity of the Omicron strain has led to speculation about its origin. The mathematical modelling platform developed for the Stockholm Paradigm (SP) indicates strongly that it has retro-colonized humans from an unidentified nonhuman mammal, likely originally infected by humans. The relationship between Omicron and all other SARS-CoV-2 variants indicates oscillations among hosts, a core part of the SP. Such oscillations result from the emergence of novel variants following colonization of new hosts, replenishing and expanding the risk space for disease emergence. The SP predicts that pathogens colonize new hosts using pre-existing capacities. Those events are thus predictable to a certain extent. Novel variants emerge after a colonization and are not predictable. This makes it imperative to take proactive measures for anticipating emerging infectious diseases (EID) and mitigating their impact. The SP suggests a policy protocol, DAMA, to accomplish this goal. DAMA comprises: DOCUMENT to detect pathogens before they emerge in new places or colonize new hosts; ASSESS to determine risk; MONITOR to detect changes in pathogen populations that increase the risk of outbreaks and ACT to prevent outbreaks when possible and minimize their impact when they occur.
Novel and previously recognized pathogens emerging in humans, crops and livestock cause costly epidemics in many parts of the planet (Brooks et al., 2019; Fauci & Morens, 2012; Morens et al., 2004; Trivellone et al., 2022). The recent COVID-19 outbreak has reached pandemic proportions, challenging understanding of the eco-evolutionary dynamics of pathogens and the way health systems cope with emerging infectious diseases (EIDs). Reported (5.54 million [World Health Organization, 2022]) and estimated (12–22 million [Adam, 2022]) deaths attributed to COVID-19 are far fewer than the pandemic caused by the 1918-Spanish Flu (influenza), which caused an estimated 50–75 million deaths worldwide (Adam, 2022; Johnson & Mueller, 2002). Nonetheless, the socioeconomic impact has been enormous, due to an absence of anticipation, inadequate preparation and uncoordinated response by healthcare systems globally.
One reason for the poor response is adherence to a paradigm of the evolutionary nature of pathogen–host associations that has clearly failed. Two fundamental shortcomings are the ideas (1) that pathogens are evolutionarily specialized for host species (host specificity), rather than for specific characteristics possessed by a given host or hosts (host resource specificity) and (2) that colonizing a new host requires the a priori evolution of novel genetic capacities. The COVID-19 pandemic in general and the Omicron emergence have highlighted the flaws in this theoretical framework.
1 REVELATIONS FROM OMICRON
The Omicron strain of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the causative agent of COVID-19 – has baffled scientists and clinicians (Venkatakrishnan et al., 2021). Phylogenetic and mutation analyses indicates that it diverged early in the outbreak and dissemination of SARS-COV-2 (Nextstrain phylogeny in 7 January 2022 – Hadfield et al., 2018). The evolution of the Spike Protein (S1) of Omicron is qualitatively and quantitatively distinct from all other lineages of SARS-COV-2 (Figure 1).
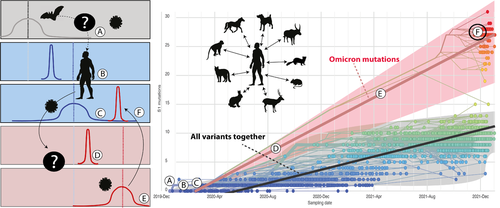
A recent op-ed piece (Kupferschmidt, 2021) presented three hypotheses posed by scientists from around the world. According to them, ‘the virus could (1) have circulated and evolved in a population with little surveillance and sequencing; (2) it could have originated in an immuno-compromised COVID-19 patient or (3) it might have evolved in a nonhuman species, from which it recently followed a trajectory of secondary colonization in people’ (hereafter H1, H2 and H3). The second option appears to be the hypothesis of choice for many, especially associated with patients with human immunodeficiency virus (Healy, 2021a, 2021b) despite recent opposing evidence (Álvarez et al., 2022). Empirical and theoretical analyses presented herein are also not in accordance with the origin of Omicron in immunodeficient humans (H1) nor in isolated human populations (H2).
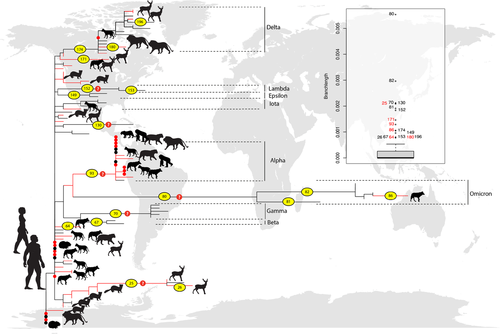
Phylogenetic comparisons and analyses of the evolution of diversity of the S1 protein in Omicron and all other variants of SARS-COV-2 show at least two distinct pathways of mutation accumulation for Omicron, especially considering the first reported Omicron sequences (Figures 1 and 2) (methodology is available in Supporting Information S1). Independent of the level of isolation of a putative human population (H1) or the vulnerability of immune-compromised human hosts (H2), the accumulation of mutations (selection) of S1 among humans would occur within or very close to the grey area indicated in Figure 1, due to common selective pressures associated with properties of the human angiotensin converting enzyme 2 (ACE2), the receptor providing the entry point for the coronavirus. Omicron variants differ from all other variants of SARS-CoV-2 in the slope of the visual regression lines (black and red lines, respectively) and in the overall number of mutations accumulated through time (Figure 1). At least one study on experimental infection (reviewed in Islam et al., 2021) indicated that ‘the rapidity of in vitro and in vivo SARS-CoV-2 selection reveals residues with functional significance during host-switching … demonstrating plasticity of viral adaptation in animal models (Bashor et al., 2021)’. Figure 2 provides further evidence for the overwhelming accumulation of mutations in the ancestral branch of the Omicron clade. Furthermore, the variability in structure of ACE2 molecules among mammals (Damas et al., 2020) also supports the third hypothesis, explaining the observed unique characteristics of early variants of the Omicron lineage in humans (black circle in Figure 1).
Empirical evidence outlined here points to a strong divergent selective pressure for the S1 in Omicron. Such a selection regime, with infection in one or more nonhuman mammalian hosts, is consistent with a high genetic and structural protein divergence from human ACE2 (Damas et al., 2020; Wei et al., 2021).
Phylogenetic reconstruction (Figure 2; Figure S1) and the proposed pathway of mutations and selection above strongly suggest that the origin of Omicron is associated with: (1) colonization of a nonhuman mammal from infected humans during the early stages of the pandemic, (2) followed by isolation and diversification under a distinct selective regime and (3) subsequent and recent re-colonization of humans (see also Sun et al., 2021; Wei et al., 2021). The wide capacity of SARS-CoV-2 to explore and exploit a diverse group of mammalian species as hosts became obvious early in the pandemic. SARS-CoV-2 emerged in humans and spread rapidly in late 2019 in an environment in which wildlife, domestic animals and humans were in unusual close contact (Figure 1a) (Boni et al., 2020; Lytras et al., 2021). Soon after, humans were reported to be a source for infections among other mammalian species (Abdelnabi et al., 2021; Islam et al., 2021; Oude Munnink et al., 2021; Shi et al., 2020). Retro-colonization of humans, evidenced early in the outbreak, has occurred with independent events in different host assemblages and multiple geographic regions (Hale et al., 2022; Kuchipudi et al., 2022; Oude Munnink et al., 2021).
Multiple events of colonization of nonhuman mammalian hosts by independent viral lineages across the evolution of SARS-CoV-2 variants and the length of the branch leading to new hosts (Figure 2) strongly indicates considerable capacity for exploring and exploiting an assemblage of host species (Kuchipudi et al., 2022; Pickering et al., 2022). Branch length (divergence under the specific substitution model used) is expected to be influenced by the genetic lagload (Smith, 1976) and/or duration of the association. Long branches may point to a potential ongoing colonization of a new host species (including retro-colonizations of other mammals and humans) (i.e., exploitation or host–range expansion) – the association exists long enough as to accumulate unique mutations under a distinct selective regime (a result of greater lagload) (see the outliers indicated in the boxplot inset of Figure 2). In contrast, short branches (composing the distribution of branch lengths in the inset boxplot of Figure 2) likely represent novel or temporary exploration of new host species, but it can suggest also a smaller lagload (in this case, close similarities of ACE2 among different host species). In summary, quantitative and qualitative changes in the genetic diversity of a lineage (i.e., amount and rate of accumulation of observed mutations) occur solely after exposure to a new selective regime. Hence, even considering the limited coverage of sequences sampled of the phylogeny in Figure 2 and the comparatively reduced sequencing of the virus from different populations and hosts species worldwide, the clade associated with Omicron (Figure 2) is fully compatible with the putative scenario suggested for the origin of the variant (Figure 1).
In fact, the analysis of the outlier branch lengths in Figure 2 (compare inset boxplot and their positions in the phylogeny) suggest additional potential cases of emergence of new genetic variants in association with host explorations and subsequent exploitations. Six out of 18 outlier branch lengths (Figure 2, inset boxplot) are linked to the colonization of new host species, and it is possible that many of the 12 remaining outliers are also associated with the same dynamics – some outlier branches may depict short length simply because the time of isolation from humans has been reduced or because they are subjected to a smaller lagload.
As suggested earlier, SARS-CoV-2 presents an immense capacity to explore and exploit a vast range of host species, most likely associated with mammals, consistent with the conservative nature of the ACE2 protein (Damas et al., 2020). While capacity of the virus is known, its presently documented host distribution is still far smaller than that predicted theoretically (Damas et al., 2020; Liu et al., 2021) and points to opportunity as a vital factor in determining the establishment of associations. Indeed, host–range changes of SARS-Cov-2 (i.e., host switching) are apparently greater among humans, nonhuman urban and peri-urban animals than game animals (Figure 2; Table S1), but the involvement of a broader assemblage of wild animals cannot be ruled out. A prevailing impression is that a reduced opportunity of encounter, and thus exposure, represents a smaller potential for risk to humans for an emergent disease (Holmes, 2022). However, as for Omicron, once the emergence occurs, new pathogens rapidly spread throughout the planet, in this case, thanks to our super-spreading abilities (see below).
2 THE COVID PANDEMIC RE-VISITED: HUMAN STEPPINGSTONES AS MEDIATORS OF PANDEMIC EXPANSION
Bats, under natural circumstances, are ecologically and behaviourally isolated from other mammals. Consequently, most of their diverse array of pathogens are known only from one or a small number of chiropteran hosts. Bats are associated with a broad range of viral pathogens, causative agents of realized or potential zoonoses (Dobson, 2005; Olival et al., 2017; Streicker & Gilbert, 2020), but direct encounters with people are infrequent (Holmes, 2022). Direct colonization of humans is rare and requires special circumstances, consistent with observations of Sarbecovirus infections that have been discovered among guano miners working in bat caves in SE Asia (Joyjinda et al., 2019; Wacharapluesadee et al., 2013). Researchers, reflecting the traditional view that host species specificity holds the key to host species switching, however, continue to be focused on chiropterans in an incomplete effort to understand the outbreak leading to the pandemic and the potential for new emergence events over time (e.g., Anthony et al., 2017; Corman et al., 2015; Dobson, 2005; Huynh et al., 2012; Joyjinda et al., 2019; Latinne et al., 2020; Leopardi et al., 2018; Li et al., 2005; McCarthy, 2021; Olival et al., 2017; Streicker & Gilbert, 2020; Valitutto et al., 2020; Wacharapluesadee et al., 2013; Wertheim et al., 2013; Young & Olival, 2016; Mallapaty, 2021; Zhou et al., 2021; Zhou & Shi, 2021).
We believe the focus should shift to a broader array of mammals. SARS-like viruses would have remained isolated among bats in the absence of humans facilitating a breakdown in ecological isolation for these assemblages. Colonization requires a steppingstone linkage from bats to humans; or from bats to other potential mammalian species that may be synchronic and sympatric; ecological fitting and sloppy fitness space bridge the nexus for capacity and opportunity (Araujo et al., 2015; Morens & Fauci, 2020). Focused circumstances, interfaces with considerable propagule pressure and viable steppingstones, are required to complete infection pathways to people (Feronato et al., 2022).
In particular, humans drove the emergence and spread of SARS-CoV-2. A shortage of pork in China due to an outbreak of ASFV in early 2019 (Brooks et al., 2022; Lytras et al., 2021; Xia et al., 2021) increased the demand for game meat in places like the live animal markets of Wuhan. This created new anthropogenic opportunity space, leading to multiple events of colonization among nonhuman mammals, some of which became steppingstone hosts for dissemination across additional mammalian hosts, including humans (Pekar et al., 2022; Worobey et al., 2022). Humans in the final quarter of 2019, in the absence of restrictions on international trade and travel, circulated the virus globally. Wherever humans established the virus, people were the primary reservoirs of infection in wildland, agricultural and urban environments (Rochman et al., 2021), serving as mediators of introduction and exposure among diverse mammalian hosts across terrestrial ecosystems (Cai & Cai, 2021; Hale et al., 2022; Kuchipudi et al., 2022; Lytras et al., 2021; Oude Munnink et al., 2021; Pickering et al., 2022; Wei et al., 2021). The animal wet markets in Wuhan and other urban centres in China and Southeast Asia provided steppingstone opportunities for SARS-like viruses to colonize humans on at least two other recent occasions – SARS-CoV in 2003 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 (Huong et al., 2020; Lytras et al., 2021; Pekar et al., 2022; Worobey et al., 2022; Xia et al., 2021; Zhou et al., 2021; Zhou & Shi, 2021).
By focusing on specific host resources, rather than host species, all mammals with Spike-ACE2 are considered potential reservoirs, and this has been borne out. Despite patterns of recombination and mutations following initial colonization, the evolutionarily conserved Spike-ACE2 protein–protein interaction has not been substantially modified (Damas et al., 2020). Mammals representing the Carnivora (Canidae, Felidae, Mustelidae and Procyonidae), Cetartiodactyla, Primates and Rodentia have been documented as competent reservoir hosts on all continents of the Northern and Southern Hemisphere, excluding Antarctica (Gryseels et al., 2021; Hale et al., 2022; Islam et al., 2021; Kuchipudi et al., 2022; Lin et al., 2021; Mahdy et al., 2020; Martins et al., 2021; Pickering et al., 2022; Sun et al., 2021; Wei et al., 2021). We must assume that unimpeded human travel in the early stages of the outbreak made SARS-CoV-2 a globally endemic pathogen with many reservoir hosts capable of producing novel variants transmitted across virtually all ecological interfaces (Figure 3) (Deng et al., 2020; Kuchipudi et al., 2022; Rochman et al., 2021) (Figure 2). SARS variants originate in isolation both in people and assemblages of domestic, semi-domestic and wild, free-ranging mammals; subsequent dissemination of initially focal variants occurs with decreased isolation and increasing connectivity (Figure 3) (Brooks et al., 2019; Feronato et al., 2022; Hoberg & Brooks, 2008; Rochman et al., 2021). The pattern described above represents a dynamic of humans facilitating opportunity as ecological super-spreaders in the origins and perpetuation of a global pandemic.
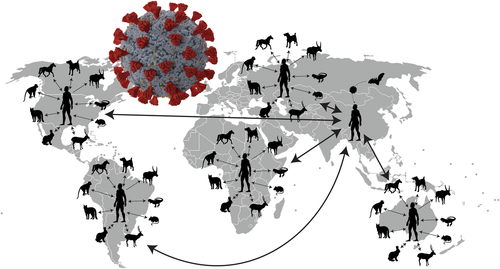
The SARS-CoV-2 pandemic is an excellent example of an evolutionary dynamic in which pre-existing genetic capacity for infecting compatible hosts, given novel exposure (opportunity) and the retention of capacity to reinfect a previous host species, is in full agreement with the recently articulated SP (Brooks et al., 2014, 2019). The SP provides an explanatory framework for both EIDs and the failure of traditional crisis–response approaches to coping with outbreaks.
3 THE STOCKHOLM PARADIGM
The SP proposes that pathogens exhibit no specificity for host species but do exhibit pronounced specificity for certain characteristics of host species (Brooks et al., 2014, 2019). The paradigm further asserts that colonization of new hosts by pathogens does not require the evolution of novel and specific genetic information related to infecting the target host. Moving into novel hosts is a matter of pathogens having the opportunity to be exposed to susceptible but previously unexposed hosts. Pathogens with specific host resource requirements may be capable of infecting a broad range of hosts if the specific resource is phylogenetically conservative and widespread (Agosta et al., 2010; Brooks & Boeger, 2019; Hoberg & Brooks, 2015; Trivellone et al., 2019). It is as simple and fundamental as recognizing that SARS-CoV-2 requires some level of compatibility between viral S1 and host's ACE2 receptors for infection (Conceicao et al., 2020; Damas et al., 2020); therefore, almost all mammals must be considered at risk for infection if exposed. A robust modelling platform has shown that EID can occur easily in this manner (Araujo et al., 2015; Braga et al., 2018; Feronato et al., 2022).
Modelling and empirical studies suggests that subsequent host changes (called oscillations in the SP [Braga et al., 2018; Janz & Nylin, 2008]) can be achieved directly or by a steppingstone dynamic, e.g., from wildlife to domestic animals to humans and vice versa (Araujo et al., 2015; Morens & Fauci, 2020), and that fast-evolving pathogens, such as viruses, retain the capacity to retro-colonize a previous host species (Feronato et al., 2022). Although initial colonization of a new host is achieved through pre-existing variation, each new host species represents a new selective regime for the colonizing pathogen population. Novel variants thus emerge after the colonization of new hosts, not before. This creates significant potential for new clades of pathogens with distinct epidemiological characteristics to emerge simply by chance. These characteristics may favour unpredictably distinct aspects in the newly acquired association, including greater or lesser virulence, or even greater transmissibility (as it appears to be the case for Omicron).
Global climate change and anthropogenic modification of ecosystems, as well as global trade and travel produce novel opportunities for EID. The risk space for EID is far larger than imagined (Brooks & Boeger, 2019; Brooks et al., 2019; Hoberg & Brooks, 2015), and is continuously replenished and expanded in time through evolution. The biosphere is replete with a growing number of evolutionary ‘accidents waiting to happen’, pathogens circulating in ecosystems and managed landscapes globally (Hoberg & Brooks, 2015). This explains why traditional approaches for coping with EID have failed. Responding only after the fact for any emergence, no matter how rapidly, is ultimately ineffective and unsustainably costly (Brooks et al., 2019, 2022; Trivellone et al., 2022). Even adequately managed EIDs may recycle in the risk space and re-emerge as distinct lineages with unique epidemiological features.
4 FINDING THEM BEFORE THEY FIND US
The SP provides some hope in dealing with the increasing risk space of EID. Both the host resources needed for colonization and the modes of transmission from host to host are generally highly specific but phylogenetically conservative. That means that we can largely predict how a given known pathogen, or a previously unknown close relative of a known pathogen, might behave when it enters a new ecosystem or encounters a susceptible host.
The DAMA (Document-Assess-Monitor-Act) protocol is a proactive policy proposal derived from the SP (Brooks et al., 2014, 2019). Document: we have increasingly fine-scale and sophisticated technology capable of documenting the diversity and distribution of potentially pathogenic micro- and macro-parasites before they announce themselves in disease outbreaks (Brooks et al., 2014, 2019; Colella et al., 2021; Hoberg & Brooks, 2015). The technological toolkit must be expanded in the context of inventories guided by the SP, focusing on potential reservoirs and environmental interfaces that facilitate persistence and transmission. DNA technology should facilitate this task, allowing us to prospect potentially dangerous pathogens in urban, peri-urban and wild animals. Also, despite general recommendations, documenting potentially zoonotic pathogens should be performed in their original hosts, prior to colonization of humans. Documenting pathogens among an expected assemblage of potential hosts or only after emergence is, in our view, profoundly dangerous. As we indicated above, once colonization occurs, the emerging pathogen will rapidly exploit and spread through human (or animal) populations. Assess: Inventories will identify more potential pathogens than we can hope to analyse effectively. We need to assess the potential risk for suites of microbes we find. This is a three-part process. (1) Phylogenetic triage: we ask two questions about each documented microbe in its phylogenetic context. First, is this a known pathogen? Second, is this a close relative of a known pathogen? If the answer to both questions is ‘no’, the microbe is deemed to be of low risk at that time. These minimum risk microbes will be accorded a status of ‘defer focused study, but archive’, recognizing their risk designation may change. If the answer to question 1 is ‘yes’, relevant health authorities should be alerted and their plans for dealing with known high-risk pathogens activated. If the answer to question 1 is ‘no’ but to question 2 is ‘yes’, the pathogen is targeted for further scrutiny. For example, we should anticipate assemblages of Betacoronavirus (including Sarbecovirus which contains SARS-CoV and SARS-CoV-2) and Alphacoronavirus pose a high risk for emergence and re-emergence (Boni et al., 2020; Cai & Cai, 2021; Damas et al., 2020; Latinne et al., 2020; Lin et al., 2021; Mallapaty, 2021; Rochman et al., 2021; Sun et al., 2021). (2) Phylogenetic assessment: using standard methods of comparative phylogenetic studies (Brooks & McLennan, 2002), we can infer the likely place of origin, the host of origin, likely transmission modes, and likely microhabitat preferences (host resource specificity) for a targeted potential pathogen. (3) Ecological assessment: understanding the population genetics and ecology of the potential pathogen, matching fitness profiles within the modelling platform to identify likely reservoir hosts and using ecological niche modelling to determine likely habitat interfaces for transmission. Monitor: Armed with the assessment, monitor each high-risk potential pathogen intensively. These efforts should focus on the interfaces between wildlands and managed habitat to find reservoir hosts that are infected but not diseased (Audy, 1958), and their transmission dynamics. The goal is to note changes in abundance, host range or geographic distributions, suggesting increased potential for outbreaks. Act: Use information from monitoring programs to determine which potential or known pathogens of high risk are already present, but not yet causing disease and which are likely to be approaching. The goal is to prevent as many outbreaks as possible and to mitigate the impact of those we cannot prevent. It is essential to archive field-based specimens for both low-risk and high-risk pathogens and their hosts, consolidating resources for biodiversity informatics linking all associated geographic, taxonomic, phylogenetic, genetic and ecological data streams (Colella et al., 2021; Dunnum et al., 2017); these are the foundations for essential baselines over space and time. Information synthesis, on databasing platforms, can drive rapid and timely availability of vital knowledge about pathogens and disease among researchers and the arena of public policy. Communications specialists subsequently translate insights from science into broad-based awareness for public security (Brooks et al., 2022; Colella et al., 2021). Information about reducing potential exposure and the early warning signs for disease outbreaks need to be widely disseminated. For example, the SP dynamic clearly demonstrates why intermittent lockdowns, rather than limiting or controlling disease outbreaks (as suggested by Keeling et al., 2021), are likely to amplify rare variants in the isolation of a lockdown, which then spread rapidly when the lockdown ends.
The SP shows how easily EID can occur in a biosphere affected by global climate change and anthropogenic modifications. We live in a world in which emerging infectious diseases will be increasingly common in crops, livestock and humans as climate change – in conjunction with globalized trade and travel – catalyses movements of hosts and pathogens, increasing the contacts between pathogens and susceptible but previously un-exposed hosts. The DAMA protocol must be at the forefront of policies aimed at reducing the socioeconomic impact of EIDs. The best chance we have for mitigating EIDs is through anticipation and protocols that enhance our abilities to find them before they find us. The SP explains why pre- and postpandemic expansion dynamics differ (Wei et al., 2021). New hosts are colonized by pathogens based on pre-existing, and thus predictable, capacities. New variants arise after a novel host is infected, and they are unpredictable. Once that happens, unsustainably expensive crisis response is our only option.
ACKNOWLEDGEMENTS
This study was partially funded by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil.
ETHICAL APPROVAL
Ethical Statement is not applicable since the study did not involve sampling animals nor humans, or questionnaires.
CONFLICT OF INTEREST
The authors report no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in Global Influenza Surveillance and Response Team at https://www.gisaid.org/. These data were derived from the following resources available in the public domain: Global Influenza Surveillance and Response Team, https://www.gisaid.org/




