Molecular detection of SARS-CoV-2 and differentiation of Omicron and Delta variant strains
Authors Wai Ning Tiffany Tsui and Vaughn Hamill contributed equally to this study.
Abstract
The SARS-CoV-2 virus is the causative agent of COVID-19 and has undergone continuous mutations throughout the pandemic. The more transmissible Omicron variant has quickly spread and is replacing the Delta variant as the most prevalent strain globally, including in the United States. A new molecular assay that can detect and differentiate both the Delta and Omicron variants was developed. A collection of 660,035 SARS-CoV-2 full- or near-full genomes, including 169,454 Delta variant and 24,202 Omicron variant strains, were used for primer and probe designs. In silico data analysis predicted an assay coverage of >99% of all strains, including >99% of the Delta and >99% of Omicron strains. The Omicron variant differential test was designed based on the Δ31-33 aa deletion in the N-gene, which is present in the original B.1.1.529 main genotype, BA.1, as well as in BA.2 and BA.3 subtypes. Therefore, the assay should detect the majority of all Omicron variant strains. Standard curves generated with human clinical samples indicated that the PCR amplification efficiencies were 104%, 90.7% and 90.4% for the Omicron, Delta, and non-Delta/non-Omicron wild-type genotypes, respectively. Correlation coefficients of the standard curves were all >0.99. The detection limit of the assay was 14.3, 32.0, and 21.5 copies per PCR reaction for Omicron, Delta, and wild-type genotypes, respectively. The assay was designed to specifically detect SAR-CoV-2 strains. Selected samples with Omicron, Delta and wild-type genotypes identified by the RT-qPCR assay were also confirmed by sequencing. The assay did not detect any animal coronavirus-positive samples that were tested. Human nasal swab samples that previously tested positive (n = 182) or negative (n = 42) for SARS-CoV-2 by the ThermoFisher TaqPath COVID-19 Combo Kit, produced the same result with the new assay. Among positive samples, 55.5% (101/182), 23.1% (42/182), and 21.4% (39/182) were identified as Omicron, Delta, and non-Omicron/non-Delta wild-type genotypes, respectively.
1 INTRODUCTION
Globally, over 349.6 million confirmed human cases, and over 5.59 million deaths have resulted from the COVID-19 pandemic (https://covid19.who.int/, January 25, 2021). In the United States, despite more than a half billion doses of vaccines that have been administered, over 70 million total cases have been confirmed (https://www.cdc.gov/, January 25, 2021). The Delta variant of SARS-CoV-2, identified in India in October, 2020, soon became the predominant variant in many countries worldwide, including the United States (Fowlkes et al., 2021; Herlihy et al., 2021; Lam-Hine et al., 2021). However, the Omicron variant emerged in November, 2021 and has quickly become the most prevalent variant in the United States and elsewhere (CDC, 2021; Del Rio et al., 2022). We recently developed an assay that detects the majority of SARS-CoV-2 strains including Delta and Omicron variants, but differentiates only the Delta variant, not the Omicron variant (Hamill et al., 2021). In the current study, we are reporting a newly developed real-time RT-PCR assay that can detect the majority of the SARS-CoV-2 strains, and differentiate both Delta and Omicron variants.
2 MATERIALS AND METHODS
2.1 Sequence analysis and assay design
Full- or near-full genomes of Omicron and Delta variants, and non-Delta/non-Omicron strains were downloaded from public databases (https://www.ncbi.nlm.nih.gov/; https://www.gisaid.org/) for analysis. A total of 660,035 genomes, including 169,454 Delta variant, 24,202 Omicron variant genomes of US origin (submitted as of December 20, 2021), and 447,359 non-Delta/non-Omicron strains were downloaded and analyzed for primer and probe designs. The 9-bp deletion, which confers Δ31–33 aa deletion in the N-gene and is unique to Omicron variants was used as a molecular target for Omicron variant detection assay design. Probes for both Omicron and the wild type were designed in the same region to allow competitional hybridization to the correct genotype template. A differential assay for the Delta variants and wild-type strains was designed in the 6-nt deletion region in the S-gene (confers Δ157–158 aa deletion) from an earlier study (Hamill et al., 2021) and was then multiplexed with the Omicron design to form a five-probe (Omicron and wild type, Delta and wild type; and 18S rRNA gene), four-channel multiplex RT-qPCR assay with both wild-type probes labelled with the same dye (Omicron: TexasRed; Delta: FAM; both wild types: VIC; and 18S rRNA gene: Cy5). The 18S rRNA gene target, used as internal control, is from a previous study (Wang et al., 2020) and amplifies well from human cells (Table 1).
| Primer/probe name | Target Genotype | Sequence (5′−3′) | Tm (°C) | Amplicon Size (bp) | Coverage (primers and probes combined) | Sources |
|---|---|---|---|---|---|---|
| Real-time PCR target primers and probes | ||||||
| SARS2-dF | Common primers for Delta (dPr) and non-Delta (wPr) probes | CCACAAAAACAACAAAAGTTGG | 59.4 | 78 for Delta, 84 for non-Delta strains | Hamill et al., 2021; strain coverage reanalyzed in this study | |
| SARS2-dR | TGAGAGACATATTCAAAAGTGCAA | 58.9 | ||||
| SARS2-dPr | Delta variant | FAM-ATAAACTCCACTTTCCA | 66.0 | 167,813/169,454 (99.0%) | ||
| SARS2-wPr | Non-Delta wild type | VIC-ATAAACTCTGAACTCACTTT | 65.0 | 426,963/447,359 (95.4%) | ||
| OmN-F | Common primers for Omicron (OmNm) and non-Omicron (OmNw) probes | GGACCCTCAGATTCAACTGG | 59.5 | 86 for Omicron, 95 for non-Omicron strains | This study | |
| OmN-R | GCAGTATTATTGGGTAAACCTTGG | 60.0 | ||||
| OmNm-Pr | Omicron variant | TexasRed-ATCGCGCCCCACCATTCT | 66.1 | 24,202/24,186 (99.9%) | ||
| OmNw-Pr | Non-Omicron wild type | VIC-CGCCCCACTGCGTTCTCC | 67.6 | 437,280/447,359 (97.7%; 99.9% with SARS2-wPr) | ||
| Real-time PCR internal control primers and probe | ||||||
| 18S-F | Human 18S ribosomal RNA gene | GGAGTATGGTTGCAAAGCTGA | 60.2 | 100 bp | 158/159 (99.4%) | Wang et al., 2020 |
| 18S-R | GGTGAGGTTTCCCGTGTTG | 61.4 | ||||
| 18S-Pr | Cy5-AAGGAATTGACGGAAGGGCA | 64.0 | ||||
| Cloning and sequencing primers | ||||||
| SARS2-cdF | Delta variant and non-Delta strains | TGGGACCAATGGTACTAAGAGG | 60.2 | 440 for Delta/446 for non-Delta strains | 9997/10,000 (99.9%) | Hamill et al., 2021 |
| SARS2-cdR | AACCCTGAGGGAGATCACG | 60.1 | ||||
| OmN-cF | Omicron variant and non-Omicron strains | CGTTGTTCGTTCTATGAAGACTTT | 58.9 | 375 for Omicron/384 for non-Omicron strains | 13,131/13,077 (99.6%) | This study |
| OmN-cR | TCATTTTACCGTCACCACCA | 59.8 | ||||
- a All quenchers followed the manufacturer's recommendations.
2.2 Clinical samples
All human clinical samples were collected by Kansas State University Lafene Health Center and transferred to Kansas State Veterinary Diagnostic Laboratory (KSVDL) for PCR testing. A total of 224 human nasal swabs that previously tested positive (n = 182) or negative (n = 42) for SARS-CoV-2 by the ThermoFisher (Carlsbad, CA) TaqPath COVID-19 Combo Kit were used for diagnostic validation of the new assay.
2.3 Positive amplification control constructions
A pair of primers, encompassing real-time PCR primer and probe regions, was used to amplify a 384 base pair (bp) region of the wild-type strain and a 375 bp region of the Omicron variant (Table 1), which then served as positive amplification controls. Amplicons with the correct size were verified by Qiagen QIAxcel (Valencia, CA, USA), then purified using the Qiagen QIAquick PCR Purification Kit. Purified PCR products were measured by a Nanodrop spectrophotometer (ThermoFisher), and directly used as positive amplification controls and also serially diluted and tested to determine the limit of detection.
2.4 RNA extraction and PCR reaction optimization
Viral RNA was extracted with MagMax Viral/Pathogen Nucleic Acid Isolation Kit (Applied Biosystems/ThermoFisher, Foster City, CA, USA) using KingFisher-96 Flex automated extraction machine (ThermoFisher). The RT-qPCR total reaction volume was 20 μl, consisting of 5 μl of 4x TaqPath 1-Step RT-qPCR Master Mix (Applied Biosystems/ThermoFisher), 5 μl of template RNA, 0.5 μM of each forward and reverse qPCR primers. To achieve similar signal intensities among each probe in the reaction, different volumes of each target probe were tested. To identify optimum annealing temperature, primers were tested with a thermal gradient ranging from 58 to 70°C. All RT-qPCR reactions were run on a Bio-Rad CFX96 Touch Real-Time PCR Detection System, and resulting cycle threshold (Ct) values of clinical samples, cloned plasmids and linear DNA were analyzed using the Bio-Rad CFX Maestro 2.2 software.
2.5 Analytical sensitivity and limit of detection
where X is the concentration in ng/μl measured by a Nanodrop spectrophotometer.
2.6 Analysis of assay's specificity
The specificity of the assay was analyzed in silico by comparing closely related coronaviruses, including SARS-CoV-1, MERS, and HKU1, 229E, NL63, and OC43 human coronavirus strains (Figure 1). Selected animal samples positive for bovine coronaviruses, canine coronavirus, porcine epidemic diarrhoea virus (PEDV), porcine deltacoronavirus (PDCoV), and porcine transmissible gastroenteritis virus (TGEV) were tested with the new SARS-CoV-2 assay (Table 5).

2.7 Sequencing confirmation of selected Omicron variant, Delta variant, and wild-type strains
Primer pairs amplifying a 384 bp region of wild type and 375 bp region of Omicron variants that were used for generating positive amplification controls were also used for Sanger sequencing confirmation of the Omicron variant genotype. Primers used in Hamill et al. (2021) were used for sequencing confirmation of the Delta variant genotype. Both primer pairs amplify their corresponding RT-qPCR targets and their flanking regions; their sequences and related information are shown in Table 1. Randomly selected samples positive for the Omicron variant, Delta variant, and the non-Omicron/non-Delta wild type, as identified by the RT-qPCR, were subjected to amplification and sequencing by the sequencing primers. Because the Delta variant genotype was sequence confirmed in a previous study (Hamill et al., 2021), only two samples were subjected to sequencing. RNA was extracted and RT-PCR amplification was performed using TaqPath 1-Step RT-qPCR Master Mix (Applied Biosystems/ThermoFisher). PCR amplicons were purified using Qiagen QIAquick PCR Purification Kit, and concentrations were measured by a Nanodrop spectrophotometer, then adjusted to optimum concentrations for Sanger sequencing. Sequencing was performed in-house using a SeqStudio (ThermoFisher) sequencing machine, and following manufacturer's instructions. The resulting raw sequencing data were trimmed and assembled using Qiagen CLC Main Workbench. Identity of assembled sequences was confirmed by comparing them to annotated Omicron variants, Delta variants, and non-Omicron/non-Delta wild-type sequences in the NCBI GenBank database.
3 RESULTS
3.1 In silico sequence analysis and primer and probe design
A vast majority of Omicron variant genomes of US origin (24,186/24,202) submitted as of December 20, 2021, contained the forward and reverse primer binding sites and the Omicron-specific probe binding site (OmNm-Pr), indicating 99.9% strain coverage for the Omicron variant genotype. Re-evaluation of the Delta variant assay designed in Hamill et al. (2021) with a larger set of data showed perfect matches of the primers and Delta-specific probe (SARS2-dPr) in 99.0% (167,813/169,454) of Delta variant genomes. Strain coverage of the wild-type assays (non-Delta and non-Omicron) corresponding to the Delta or Omicron target locations were 95.4% (426,963/447,359) and 97.7% (437,280/447,359), respectively, with a combined strain coverage of 99.9% (446,950/447,359) (Table 1).
3.2 PCR conditions and optimization
The RT-qPCR total reaction volume was 20 μl, consisting of 5 μl of template RNA, 0.5 μM of each forward and reverse qPCR primers, and optimized with 0.375 μM of Omicron probe (OmNm-Pr), 0.5 μM non-Omicron probe (OmNw-Pr), 0.25 μM of SARS2-delta probe (SARS2-dPr), 0.5 μM of SARS2 wild type (SARS2-wPr), 0.5 μM 18S probe, and 5 μl of 4x TaqPath 1-Step RT-qPCR Master Mix (Applied Biosystems/ThermoFisher). The optimum annealing temperature for the assay was 60.4°C as determined by a thermal gradient test ranging from 58 to 70°C. Accordingly, the thermocycling parameters used for the following experiments were set with an RT reaction at 48°C for 10 min, then an inactivation and denaturation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 20 s and annealing/extension at 60°C for 40 s.
3.3 Standard curve analysis of genotype-confirmed plasmids and purified DNA products
Genotypes of cloned plasmids containing Delta and non-Delta genotypes, and purified linear DNA products of Omicron and non-Omicron genotypes were confirmed by testing with the prototype of this RT-qPCR assay, and further confirmed by DNA sequencing prior to standard curve testing. Standard curves indicated that PCR amplification efficiencies for Omicron, Delta, and non-Delta/non-Omicron wild types were 99.3%, 97.8%, and 96.6%, respectively. Correlation coefficients (R2) for all targets were >0.99 (Figure 2a).
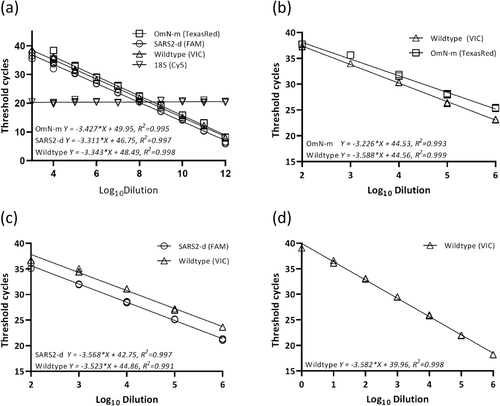
3.4 Limit of detection determination and copy number analysis
Standard curve analysis of serially diluted templates showed end-point Ct values between 36 and 37 for each target. Based on the concentration and size of the plasmids or purified DNA products, and the dilution factor that they reached, the calculated copy numbers per PCR reaction were 14.5, 32.0, and 21.5 copies for Omicron, Delta, and the wild-type genotypes, respectively (Table 2).
| SARS-CoV-2 genotypes | Omicron | Delta | Wild type |
|---|---|---|---|
| Concentrations (ng/μl) | 13.0 | 312.7 | 189.7 |
| Average endpoint Ct | 36.3 | 36.5 | 37.0 |
| Endpoint copy number (per μl) | 2.9 | 6.4 | 4.3 |
| Endpoint copy number (per PCR reaction) | 14.5 | 32 | 21.5 |
3.5 Standard curve analysis using clinical samples
PCR amplification efficiency and correlation coefficient (R2) generated by standard curve analyses with 10-fold dilutions of clinical samples containing different variants and the wild type of SARS-CoV-2 are summarized in Table 3. These samples were chosen due to their high viral concentrations, allowing for testing in a wider dynamic range of detections. For the Omicron genotype, the PCR amplification efficiency was 104.2% with a correlation coefficient (R2) of 0.992 (Figure 2b); analysis with the Delta variant revealed a PCR amplification efficiency of 90.7% with R2 of 0.997 (Figure 2c); for the non-Omicron/non-Delta wild-type strain, the PCR amplification efficiency was 90.4% and the R2 was 0.999 (Figure 2d).
| RT-qPCR assay | Plasmid or linear DNA | Diagnostic sample | |||
|---|---|---|---|---|---|
| E | R2 | E | R2 | ||
| Omicron | Multiplex | 99.3% | 0.998 | 104.2% | 0.992 |
| Singular | 100.7% | 0.990 | 90.10 | 0.999 | |
| Delta | Multiplex | 97.8% | 0.997 | 90.7% | 0.997 |
| Singular | 103.0% | 0.993 | 91.0% | 0.997 | |
| Wild type | Multiplex | 96.6% | 0.995 | 90.4% | 0.999 |
| Singular | 102.9% | 0.993 | 90.2% | 0.998 | |
- Note: E: PCR amplification efficiency; R2: correlation coefficient.
3.6 Specificity of the assay
3.6.1 In silico analysis of closely related human coronaviruses
Results from an in silico analysis of five sequences each from the Omicron and Delta variants, and non-Omicron/non-Delta wild type of SARS-CoV-2, SARS-CoV-1, MERS, and one sequence each for HKU1, 229E, NL63, and OC43 human coronavirus strains indicated that the newly designed SARS-CoV-2 assay does not match any of above-mentioned strains, except for SARS-CoV-1 that had some sequence homology. The 9-bp deletion corresponding to the Δ31-33 aa deletion in the N-gene in the Omicron variant probe is very specific and had no sequence match to SARS-CoV-1 sequences. The non-Omicron wild-type probe had some sequence homology to the N-gene of SARS-CoV-1, however, there are 2 bp differences near the middle and 1 bp difference at the end of the probe, which still distinguish it from SARS-CoV-1. Additionally, the common forward primer for both Omicron and non-Omicron genotypes ended with a specific nucleotide on the 3′ end that differentiated it from the same nucleotide site on SARS-CoV-1, which should ensure specific amplification from SARS-CoV-2 strains. Otherwise, the two newly designed SARS-CoV-2 targets are highly specific to their respective genotypes (Figure 1).
3.6.2 Sequencing confirmation of selected clinical samples of Omicron and Delta variants, and wild-type strains
Sequencing results of 13 Omicron variant strains, eight wild-type strains and two Delta variant strains confirmed the genotypes identified by the RT-qPCR assay. The best matching NCBI accessions and their corresponding percentage of homology are listed in Table 4.
| Genotype by RT-qPCR | Sample ID | Best match in NCBI | |
|---|---|---|---|
| % Identity | NCBI Accession | ||
| Omicron variant | 1 | 100% | OM185476.1 |
| 2 | 100% | OM185219.1 | |
| 3 | 100% | OM185219.1 | |
| 4 | 100% | OM185219.1 | |
| 5 | 100% | OM185219.1 | |
| 6 | 100% | OM212328.1 | |
| 7 | 100% | OM212328.1 | |
| 8 | 100% | OM212328.1 | |
| 9 | 99.7% | OM212328.1 | |
| 10 | 100% | OM212328.1 | |
| 11 | 100% | OM212328.1 | |
| 12 | 100% | OM212328.1 | |
| 13 | 99.7% | OM212328.1 | |
| Wild type | 1 | 100% | OM211960.1 |
| 2 | 100% | OM212002.1 | |
| 3 | 100% | OL706961.1 | |
| 4 | 100% | OM212002.1 | |
| 5 | 100% | OK435535.1 | |
| 6 | 100% | OK653431.1 | |
| 7 | 100% | OL337112.1 | |
| 8 | 100% | OM211960.1 | |
| Delta variant | 1 | 100% | OM139389.1 |
| 2 | 100% | OM139389.1 | |
3.6.3 Animal coronavirus testing
All animal samples in our current collection, that previously tested positive for multiple species of coronavirus, tested negative by the new SARS-CoV-2 RT-qPCR, indicating no cross-detection of animal coronaviruses (Table 5).
| Animal coronavirus samples | Ct of animal assays | Ct of SARS-CoV-2 assay | |
|---|---|---|---|
| Bovine | Enteric | 18.5 | 0.0 |
| Enteric | 20.2 | 0.0 | |
| Enteric | 13.2 | 0.0 | |
| Enteric | 19.5 | 0.0 | |
| Respiratory | 23.0 | 0.0 | |
| Canine | Enteric | 17.1 | 0.0 |
| Enteric | 24.1 | 0.0 | |
| Enteric | 22.4 | 0.0 | |
| Enteric | 22.7 | 0.0 | |
| Enteric | 17.2 | 0.0 | |
| PEDV | Enteric | 23.2 | 0.0 |
| Enteric | 30.0 | 0.0 | |
| Enteric | 29.7 | 0.0 | |
| Enteric | 15.3 | 0.0 | |
| Enteric | 19.1 | 0.0 | |
| PDCoV | Enteric | 17.2 | 0.0 |
| Enteric | 17.0 | 0.0 | |
| Enteric | 25.3 | 0.0 | |
| Enteric | 17.0 | 0.0 | |
| Enteric | 15.4 | 0.0 | |
| TGEV | Enteric | 13.3 | 0.0 |
- Abbreviations: PDCoV, porcine deltacoronavirus; PEDV, porcine epidemic diarrhoea virus; TGEV, transmissible gastroenteritis virus (porcine).
3.7 Diagnostic validation using human clinical samples
All 182 human clinical samples used in this study that previously tested positive for SARS-CoV-2 by the ThermoFisher TaqPath COVID-19 Combo Kit were also tested positive by our newly developed RT-qPCR assay. Out of the 182 clinical samples, 39 were identified as non-Omicron/non-Delta wild-type strains, while 42 and 101 tested positive for the Delta and Omicron variants, respectively (Table 6). There was no signal detected from all 101 Omicron variant samples for the S-gene target when tested by the ThermoFisher TaqPath COVID-19 Combo Kit. This phenomenon is referred to as “S-gene drop out,” and is considered an indication of detection of Omicron variant under the current pandemic situation. The 42 SARS-COV-2 negative samples tested by the ThermoFisher TaqPath COVID-19 Combo Kit were also negative by the new assay (data not shown).
| Genotype | Sample ID | Delta | Wild-type | Omicron | 18S rRNA |
|---|---|---|---|---|---|
| Non-Delta, non-Omicron wild type | 1 | 0.00 | 19.66 | 0.00 | 13.50 |
| 2 | 0.00 | 23.91 | 0.00 | 14.27 | |
| 3 | 0.00 | 21.28 | 0.00 | 15.86 | |
| 4 | 0.00 | 23.22 | 0.00 | 20.84 | |
| 5 | 0.00 | 18.49 | 0.00 | 15.83 | |
| 6 | 0.00 | 15.60 | 0.00 | 13.95 | |
| 7 | 0.00 | 24.45 | 0.00 | 25.60 | |
| 8 | 0.00 | 21.46 | 0.00 | 15.04 | |
| 9 | 0.00 | 18.80 | 0.00 | 14.33 | |
| 10 | 0.00 | 24.86 | 0.00 | 15.16 | |
| 11 | 0.00 | 16.26 | 0.00 | 14.16 | |
| 12 | 0.00 | 28.70 | 0.00 | 24.58 | |
| 13 | 0.00 | 25.56 | 0.00 | 22.55 | |
| 14 | 0.00 | 30.05 | 0.00 | 24.11 | |
| 15 | 0.00 | 23.03 | 0.00 | 24.39 | |
| 16 | 0.00 | 18.87 | 0.00 | 19.73 | |
| 17 | 0.00 | 29.28 | 0.00 | 15.29 | |
| 18 | 0.00 | 29.60 | 0.00 | 11.06 | |
| 19 | 0.00 | 35.80 | 0.00 | 26.99 | |
| 20 | 0.00 | 30.15 | 0.00 | 15.42 | |
| 21 | 0.00 | 20.63 | 0.00 | 14.05 | |
| 22 | 0.00 | 32.04 | 0.00 | 25.78 | |
| 23 | 0.00 | 17.30 | 0.00 | 19.60 | |
| 24 | 0.00 | 18.09 | 0.00 | 19.22 | |
| 25 | 0.00 | 21.48 | 0.00 | 13.43 | |
| 26 | 0.00 | 21.20 | 0.00 | 16.39 | |
| 27 | 0.00 | 26.32 | 0.00 | 16.14 | |
| 28 | 0.00 | 28.54 | 0.00 | 20.68 | |
| 29 | 0.00 | 19.64 | 0.00 | 14.15 | |
| 30 | 0.00 | 26.70 | 0.00 | 17.18 | |
| 31 | 0.00 | 27.45 | 0.00 | 16.44 | |
| 32 | 0.00 | 28.63 | 0.00 | 23.67 | |
| 33 | 0.00 | 28.08 | 0.00 | 16.72 | |
| 34 | 0.00 | 28.76 | 0.00 | 23.98 | |
| 35 | 0.00 | 35.51 | 0.00 | 17.09 | |
| 36 | 0.00 | 36.90 | 0.00 | 15.10 | |
| 37 | 0.00 | 36.87 | 0.00 | 17.13 | |
| 38 | 0.00 | 36.20 | 0.00 | 22.41 | |
| 39 | 0.00 | 36.17 | 0.00 | 20.32 | |
| Delta variant | 1 | 25.16 | 29.48 | 0.00 | 14.99 |
| 2 | 25.28 | 30.35 | 0.00 | 17.90 | |
| 3 | 21.37 | 26.11 | 0.00 | 14.38 | |
| 4 | 26.72 | 30.36 | 0.00 | 22.08 | |
| 5 | 22.14 | 26.49 | 0.00 | 20.70 | |
| 6 | 25.45 | 29.55 | 0.00 | 15.71 | |
| 7 | 21.66 | 25.39 | 0.00 | 18.80 | |
| 8 | 30.05 | 34.42 | 0.00 | 19.14 | |
| 9 | 29.91 | 35.50 | 0.00 | 15.60 | |
| 10 | 18.84 | 24.21 | 0.00 | 16.15 | |
| 11 | 25.24 | 30.41 | 0.00 | 19.50 | |
| 12 | 26.67 | 31.23 | 0.00 | 17.15 | |
| 13 | 23.04 | 28.97 | 0.00 | 17.90 | |
| 14 | 19.45 | 25.02 | 0.00 | 23.24 | |
| 15 | 26.14 | 31.68 | 0.00 | 17.59 | |
| 16 | 21.34 | 25.52 | 0.00 | 19.61 | |
| 17 | 20.07 | 24.72 | 0.00 | 18.57 | |
| 18 | 28.23 | 33.87 | 0.00 | 16.77 | |
| 19 | 22.60 | 27.25 | 0.00 | 19.32 | |
| 20 | 19.17 | 24.08 | 0.00 | 20.17 | |
| 21 | 30.41 | 36.26 | 0.00 | 15.98 | |
| 22 | 26.42 | 31.00 | 0.00 | 20.64 | |
| 23 | 28.67 | 32.90 | 0.00 | 21.35 | |
| 24 | 20.35 | 25.19 | 0.00 | 22.12 | |
| 25 | 24.09 | 27.65 | 0.00 | 29.76 | |
| 26 | 18.98 | 23.76 | 0.00 | 21.23 | |
| 27 | 20.35 | 25.35 | 0.00 | 22.21 | |
| 28 | 25.04 | 30.47 | 0.00 | 14.77 | |
| 29 | 24.94 | 30.06 | 0.00 | 14.10 | |
| 30 | 19.96 | 24.45 | 0.00 | 16.58 | |
| 31 | 19.68 | 24.70 | 0.00 | 20.15 | |
| 32 | 18.52 | 23.59 | 0.00 | 13.93 | |
| 33 | 21.26 | 24.83 | 0.00 | 14.86 | |
| 34 | 25.71 | 30.07 | 0.00 | 17.60 | |
| 35 | 20.55 | 24.44 | 0.00 | 14.44 | |
| 36 | 20.10 | 25.12 | 0.00 | 16.18 | |
| 37 | 22.75 | 27.81 | 0.00 | 15.97 | |
| 38 | 18.96 | 23.05 | 0.00 | 16.38 | |
| 39 | 18.50 | 21.25 | 0.00 | 16.49 | |
| 40 | 17.64 | 22.30 | 0.00 | 18.25 | |
| 41 | 21.39 | 23.86 | 0.00 | 16.41 | |
| 42 | 25.71 | 28.39 | 0.00 | 19.68 | |
| Omicron variant | 1 | 0.00 | 32.96 | 37.06 | 20.37 |
| 2 | 0.00 | 21.05 | 23.50 | 16.24 | |
| 3 | 0.00 | 21.84 | 23.51 | 21.09 | |
| 4 | 0.00 | 31.65 | 34.43 | 22.01 | |
| 5 | 0.00 | 30.98 | 33.52 | 17.18 | |
| 6 | 0.00 | 23.45 | 25.36 | 20.12 | |
| 7 | 0.00 | 22.98 | 25.54 | 19.87 | |
| 8 | 0.00 | 29.48 | 31.17 | 14.50 | |
| 9 | 0.00 | 33.90 | 36.07 | 22.08 | |
| 10 | 0.00 | 30.92 | 33.95 | 16.62 | |
| 11 | 0.00 | 22.00 | 24.30 | 16.30 | |
| 12 | 0.00 | 22.11 | 24.40 | 17.39 | |
| 13 | 0.00 | 24.25 | 26.41 | 23.99 | |
| 14 | 0.00 | 31.65 | 34.54 | 16.67 | |
| 15 | 0.00 | 29.42 | 32.56 | 16.35 | |
| 16 | 0.00 | 31.11 | 33.84 | 20.99 | |
| 17 | 0.00 | 21.11 | 23.50 | 15.36 | |
| 18 | 0.00 | 22.02 | 24.03 | 18.94 | |
| 19 | 0.00 | 34.84 | 36.63 | 16.18 | |
| 20 | 0.00 | 24.47 | 26.37 | 18.67 | |
| 21 | 0.00 | 20.22 | 22.19 | 19.24 | |
| 22 | 0.00 | 22.59 | 24.20 | 18.12 | |
| 23 | 0.00 | 32.59 | 35.64 | 14.04 | |
| 24 | 0.00 | 24.57 | 26.79 | 14.78 | |
| 25 | 0.00 | 25.48 | 28.04 | 17.44 | |
| 26 | 0.00 | 27.89 | 29.14 | 18.56 | |
| 27 | 0.00 | 27.42 | 29.81 | 16.15 | |
| 28 | 0.00 | 31.04 | 32.08 | 16.47 | |
| 29 | 0.00 | 21.24 | 23.19 | 16.43 | |
| 30 | 0.00 | 28.17 | 31.30 | 21.94 | |
| 31 | 0.00 | 25.27 | 28.45 | 18.02 | |
| 32 | 0.00 | 31.25 | 34.45 | 15.78 | |
| 33 | 0.00 | 33.78 | 35.35 | 24.33 | |
| 34 | 0.00 | 32.63 | 35.52 | 23.36 | |
| 35 | 0.00 | 30.80 | 33.21 | 17.87 | |
| 36 | 0.00 | 27.13 | 30.02 | 19.35 | |
| 37 | 0.00 | 33.16 | 36.87 | 18.29 | |
| 38 | 0.00 | 27.16 | 29.37 | 20.20 | |
| 39 | 0.00 | 23.97 | 26.06 | 17.49 | |
| 40 | 0.00 | 25.79 | 28.21 | 20.02 | |
| 41 | 0.00 | 25.38 | 28.26 | 18.71 | |
| 42 | 0.00 | 32.82 | 36.94 | 17.07 | |
| 43 | 0.00 | 21.01 | 22.91 | 18.62 | |
| 44 | 0.00 | 31.29 | 33.48 | 17.20 | |
| 45 | 0.00 | 22.49 | 25.01 | 15.03 | |
| 46 | 0.00 | 31.08 | 34.23 | 19.11 | |
| 47 | 0.00 | 23.45 | 25.98 | 20.99 | |
| 48 | 0.00 | 21.82 | 23.98 | 16.32 | |
| 49 | 0.00 | 23.20 | 26.11 | 16.58 | |
| 50 | 0.00 | 23.75 | 26.47 | 22.71 | |
| 51 | 0.00 | 18.18 | 20.59 | 14.58 | |
| 52 | 0.00 | 26.29 | 29.24 | 21.24 | |
| 53 | 0.00 | 21.58 | 25.09 | 15.16 | |
| 54 | 0.00 | 18.41 | 18.91 | 16.60 | |
| 55 | 0.00 | 23.38 | 25.22 | 22.77 | |
| 56 | 0.00 | 23.72 | 26.10 | 19.19 | |
| 57 | 0.00 | 23.17 | 25.97 | 17.17 | |
| 58 | 0.00 | 22.50 | 24.93 | 20.50 | |
| 59 | 0.00 | 28.24 | 30.83 | 24.58 | |
| 60 | 0.00 | 34.02 | 35.62 | 18.11 | |
| 61 | 0.00 | 35.22 | 36.85 | 17.27 | |
| 62 | 0.00 | 22.02 | 23.18 | 17.23 | |
| 63 | 0.00 | 26.01 | 28.31 | 17.85 | |
| 64 | 0.00 | 27.16 | 29.55 | 19.12 | |
| 65 | 0.00 | 28.00 | 31.33 | 17.90 | |
| 66 | 0.00 | 22.07 | 24.35 | 20.48 | |
| 67 | 0.00 | 32.55 | 36.72 | 22.08 | |
| 68 | 0.00 | 31.53 | 31.22 | 16.91 | |
| 69 | 0.00 | 33.13 | 34.14 | 19.99 | |
| 70 | 0.00 | 32.33 | 34.42 | 16.46 | |
| 71 | 0.00 | 31.25 | 33.42 | 19.99 | |
| 72 | 0.00 | 28.20 | 30.57 | 19.34 | |
| 73 | 0.00 | 24.64 | 26.83 | 17.47 | |
| 74 | 0.00 | 25.61 | 27.54 | 15.60 | |
| 75 | 0.00 | 32.22 | 33.89 | 16.88 | |
| 76 | 0.00 | 27.30 | 29.90 | 20.69 | |
| 77 | 0.00 | 29.84 | 33.46 | 27.92 | |
| 78 | 0.00 | 25.44 | 27.16 | 17.18 | |
| 79 | 0.00 | 27.15 | 29.37 | 19.34 | |
| 80 | 0.00 | 24.90 | 26.53 | 25.37 | |
| 81 | 0.00 | 24.23 | 26.54 | 22.52 | |
| 82 | 0.00 | 22.46 | 24.82 | 16.96 | |
| 83 | 0.00 | 33.76 | 35.46 | 18.27 | |
| 84 | 0.00 | 29.49 | 33.31 | 18.34 | |
| 85 | 0.00 | 22.57 | 24.69 | 22.65 | |
| 86 | 0.00 | 33.25 | 36.38 | 20.53 | |
| 87 | 0.00 | 31.58 | 34.22 | 18.01 | |
| 88 | 0.00 | 29.24 | 32.00 | 20.18 | |
| 89 | 0.00 | 30.39 | 32.82 | 18.15 | |
| 90 | 0.00 | 24.25 | 26.39 | 23.28 | |
| 91 | 0.00 | 32.73 | 36.26 | 18.11 | |
| 92 | 0.00 | 28.45 | 31.14 | 18.24 | |
| 93 | 0.00 | 20.19 | 22.59 | 17.65 | |
| 94 | 0.00 | 24.44 | 26.93 | 18.05 | |
| 95 | 0.00 | 28.49 | 31.51 | 17.58 | |
| 96 | 0.00 | 23.99 | 26.03 | 19.59 | |
| 97 | 0.00 | 24.56 | 27.21 | 19.62 | |
| 98 | 0.00 | 25.18 | 28.02 | 20.77 | |
| 99 | 0.00 | 30.68 | 33.28 | 17.31 | |
| 100 | 0.00 | 25.55 | 28.14 | 21.24 | |
| 101 | 0.00 | 24.37 | 27.08 | 19.71 | |
| No template control (NTC) | 0.00 | 0.00 | 0.00 | – | |
| Positive amplification control (PAC) | 26.12 | 24.25 | 27.97 | – | |
4 DISCUSSION
The emergence of multiple variants of concern (VOC), including the Omicron variant, has introduced additional challenges to SARS-CoV-2 diagnostics and disease management (Dejnirattisai et al., 2021; Safarchi et al., 2021; Zimmerman et al., 2021). The most prevalent VOC circulating in the United States has changed from the previously dominant Delta variant to the Omicron variant in a matter of a 1-month period (Lee, 2021; Li et al., 2021) (https://www.cdc.gov/). Some reports have indicated that infection with the Omicron variant may increase neutralizing immunity against the Delta variant (Khan et al., 2021), while other reports have demonstrated the Omicron variant has partially reduced the effectiveness of antibody protections induced by vaccinations (Doria-Rose et al., 2021; Edara et al., 2021; Farinholt et al., 2021; Garcia-Beltran et al., 2021). Detection of SARS-CoV-2 variants predominantly relies on Sanger sequencing or next-generation sequencing (Cele et al., 2021; Cherian et al., 2021; Zimmerman et al., 2021), which are labour intensive and require a long turn-around time to results. According to CDC tracking data, on January 1, 2022, 95.4% of SARS-CoV-2 human infections in the United States are from the Omicron variant, while the remaining 4.6% are from the Delta variant. Although various detection assays have been developed, diagnostic challenges remain, especially from newly emerged variant strains (Thomas et al., 2021). An accurate and sensitive assay for the detection and differentiation of Delta and Omicron variants, with a rapid turnaround time, can serve as an important public health tool for variant tracing and epidemiological investigations.
The Omicron variant has noticeably increased transmissibility, largely owed to the number of additional mutations it gained (Papanikolaou et al., 2022; Petersen et al., 2022; Syed et al., 2022). These include a large number of point mutations and deletions in the S-gene, as well as in N-gene and other parts of the genome (European Centre for Disease Prevention and Control, 2021a, 2021b). Molecular assays targeting multi-nucleotide deletions are generally more specific than assays based on a single-nucleotide mutation (Hamill et al., 2021). For the Omicron variant, there are several multi-nucleotide deletions in the S-gene, NSP6, and the N-gene. The N-gene in coronaviruses, including in SARS-CoV-2, is generally considered more conserved than the S-gene (Fang & Shi, 2022). For that reason, we choose the 9-bp deletion corresponding to the Δ31-33 aa deletion in the N-gene as the detection target. An in silico analysis predicted that the assay should detect 99.9% of 24,202 Omicron sequences of US origin that were submitted, as of December 20, 2021. Although phylogenetically, Omicron variants belong to clades 21K and 21L, both are part of a larger clade, 21 M (https://nextstrain.org/ncov/gisaid/global), which is based on mutations they mutually possess; the variant is further divided into BA.1 and BA.2 subtypes, and some reports have also proposed the BA.3 subtype. Current data indicate that all three subtypes contain the Δ31-33 aa deletion in the N-gene (Hasetline, 2022; https://covariants.org/variants/21L.Omicron), therefore the assay should provide detection for each of these subtypes. Combined with the Delta variant PCR that we recently developed (Hamill et al., 2021), these assays can detect and differentiate >99% of both Delta and Omicron variant strains.
In differential genotyping PCR assays that target on a few bp deletion or a single nucleotide polymorphism site, better genotyping results can be obtained via competitional hybridization, a binding property made possible when a wild-type probe is designed at the same site of the mutant genotype probe (Hamill et al., 2021). The non-Omicron wild-type assay has a strain coverage of 97.7%, but together with the non-Delta wild-type test, and assay detected 99.9% of sequences analyzed (446,950/ 447,359), indicating a very high coverage for non-Delta/non-Omicron wild-types strains.
Unlike the Delta variant assay, which had no matching probe sequences to closely related human coronaviruses, in this Omicron assay, the non-Omicron wild-type probe had some sequence similarity to SARS-CoV-1 strains associated with an outbreak in Asia nearly 20 years ago. However, the probe had a 2-bp difference in the middle and a 1-bp difference at the 3′ end. Furthermore, the forward primer was specifically designed to land at a different nucleotide, which together should provide specific amplification and detection of SARS-CoV-2 sequences only (Figure 1). With nearly 100% circulating strains in the United States and many other countries or regions identified as Omicron or Delta variants, the similarity of the non-Omicron wild-type test to SARS-CoV-1 N-gene sequences should not affect the detection accuracy of the assay.
We hope the high transmissibility and relatively low virulent nature of the Omicron variant that is in predominant circulation will lead to the end of the pandemic. We also understand that the genomes of viruses are continuously mutating. We will continue to monitor for changes to SARS-CoV-2 genomes, and keep our assay up-to-date in order to detect the majority of contemporary SARS-CoV-2 strains.
ACKNOWLEDGMENT
This study was supported by Kansas State Veterinary Diagnostic Laboratory, Kansas State University.
CONFLICT OF INTEREST
All authors declare no conflict of interest.
ETHICAL STATEMENT
All human nasal swab samples were collected by Lafene Health Center, Kansas State University, and submitted to KSVDL Public Health Laboratory for diagnosis under CLIA certification # 17D0648239. Research activities of this study are also authorized under Kansas State University IBC # 1619 (renewal of IBC # 1322).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in The GenBank at https://www.ncbi.nlm.nih.gov/. These data were derived from the following resources available in the public domain:
The GenBank, https://www.ncbi.nlm.nih.gov/




