How do genetic relatedness and spatial proximity shape African swine fever infections in wild boar?
Abstract
The importance of social and spatial structuring of wildlife populations for disease spread, though widely recognized, is still poorly understood in many host-pathogen systems. In particular, system-specific kin relationships among hosts can create contact heterogeneities and differential disease transmission rates. Here, we investigate how distance-dependent infection risk is influenced by genetic relatedness in a novel host-pathogen system: wild boar (Sus scrofa) and African swine fever (ASF). We hypothesized that infection risk would correlate positively with proximity and relatedness to ASF-infected individuals but expected those relationships to weaken with the distance between individuals due to decay in contact rates and genetic similarity. We genotyped 323 wild boar samples (243 ASF-negative and 80 ASF-positive) collected in north-eastern Poland in 2014–2016 and modelled the effects of geographic distance, genetic relatedness and ASF virus transmission mode (direct or carcass-based) on the probability of ASF infection. Infection risk was positively associated with spatial proximity and genetic relatedness to infected individuals with generally stronger effect of distance. In the high-contact zone (0–2 km), infection risk was shaped by the presence of infected individuals rather than by relatedness to them. In the medium-contact zone (2–5 km), infection risk decreased but was still associated with relatedness and paired infections were more frequent among relatives. At farther distances, infection risk further declined with relatedness and proximity to positive individuals, and was 60% lower among un-related individuals in the no-contact zone (33% in10–20 km) compared among relatives in the high-contact zone (93% in 0–2 km). Transmission mode influenced the relationship between proximity or relatedness and infection risk. Our results indicate that the presence of nearby infected individuals is most important for shaping ASF infection rates through carcass-based transmission, while relatedness plays an important role in shaping transmission rates between live animals.
1 INTRODUCTION
Spatial and social behaviour of the hosts plays a major role in shaping patterns of pathogen spread in animal populations (Albery et al., 2021; Altizer et al., 2003; Dougherty et al., 2018; Sah et al., 2018). Host movements define the spatial dimension of pathogen transmission while social structure determines the encounter rate between infected and susceptible individuals. In social systems with stable group membership, individual contacts and pathogen transmission occur mainly within social groups, potentially limiting the speed of disease spread (Pepin et al., 2020). This type of social structure is typically based on familial (e.g., matrilineal) groups where social interactions and disease transmission rates are correlated with genetic relatedness (Benton et al., 2016; Carter et al., 2013; Grear et al., 2010). In contrast, in fission-fusion societies with dynamic group membership, contact rates and disease transmission tend to be independent of relatedness (Hirsch et al., 2013; Mejía-Salazar et al., 2017; Vander Wal et al., 2012). The social system can, thus, affect the rate and mechanisms of pathogen transmission (Altizer et al., 2003; Sah et al., 2018). Contact heterogeneity due to social structure may be particularly relevant for disease transmission at a local scale, where groups are already in spatial proximity allowing for contact. Understanding the relative contribution of spatial and social processes in disease transmission at such scales is important for modelling and managing wildlife diseases (Dougherty et al., 2018; Pepin et al., 2021). However, investigating the role of host social and spatial behaviour in disease transmission is challenging, ideally requiring simultaneous host contact, movement and infection data and studies addressing this issue with relevant data are limited. Additionally, host social system and pathogen characteristics (e.g., infectiousness, transmission mode, lethality) interact to produce varying spatial infection patterns and epidemiological outcomes (Pepin & VerCauteren, 2016; VanderWaal & Ezenwa, 2016). Genetic structure of the population reflects long-term gene flow and social structure (Rossiter et al., 2012). Because genetic structuring and transmission of a fast-paced pathogen, such as ASF virus, operate at different timescales, we were not interested in the interaction of these two processes. Instead, we use genetic data to better understand social structuring, at a timescale compatible with pathogen transmission, and test whether the existing social structure has any role in shaping the spatial dynamics of the disease.
African swine fever (ASF) is a contagious viral disease with both direct and environmental, mainly carcass-based, transmission in Eurasian wild boar Sus scrofa which is the sole wild reservoir of the disease in Eastern Europe (Chenais et al., 2018). The ASF virus strain currently circulating in Eastern Europe (genotype II) is highly virulent, causing lethality approaching 100% within 1–3 weeks post-infection (Blome et al., 2013) and the disease causes high mortality in susceptible host populations which can reduce wild boar numbers by as much as 90% during the initial phase of an outbreak (Morelle et al., 2020). ASF virus is resistant to environmental factors and can remain active in contaminated tissues from several weeks to months (Fischer et al., 2020). Transmission through infected carcasses (i.e., indirect) has been estimated to account for about a half of all infections and contribute to long-term persistence of the disease particularly at low host densities (Pepin et al., 2020). Because diseased animals tend to die locally (i.e., within their home range), they will be a source of infections mainly for the most proximate individuals from their own or neighbouring social groups. However, transmission rates via indirect and direct (i.e., through social interactions) routes will probably differ due to varying contact dynamics and long availability of infectious carcasses (Cukor et al., 2020; Probst et al., 2020; Probst et al., 2017). Thus, carcass-based transmission interacts with direct transmission to shape local infection patterns (Lange & Thulke, 2016). The spread of infectious diseases occurs over multiple spatial scales (Riley, 2007). On the landscape level, ASF prevalence and spread correlate positively with wild boar density (Nurmoja et al., 2017; Podgórski et al., 2020), proportion of forest cover (Dellicour et al., 2020; Podgórski et al., 2020) and negatively with distance to previous cases (Podgórski et al., 2020) and physical barriers to wild boar movement (Dellicour et al., 2020). At fine scales, ASF transmission is likely influenced by a combination of social interactions, movements and spatial distribution of individuals (Pepin et al., 2021).
Wild boar social structure is based on cohesive, matrilineal social units (Gabor et al., 1999; Kaminski et al., 2005; Podgórski, Lusseau et al., 2014). Contact rates are strongly structured socially and spatially. The rate of inter-group interactions is relatively low and declines sharply with the distance between the groups. The highest contact rates are between immediately adjacent groups (0–2 km) and drop to very low levels at a distance as close as 4 km (Pepin et al., 2016; Podgórski et al., 2018; Yang et al., 2020). Such type of social structure is not conducive to rapid spread of infectious diseases (Pepin & VerCauteren, 2016). Social behaviour of wild boar, next to its sedentary lifestyle, is probably one of the factors responsible for slow natural spread of ASF in wild boar populations. The velocity of spread in the study area was 1.5 km/month (Podgórski & Śmietanka, 2018) but the estimated speeds may vary widely within the European Union, ranging from 0.6 to 54 km/month (Iglesias et al., 2019). While wild boar movements were shown to be poor predictors of ASF spread (Podgórski & Śmietanka, 2018), the role of genetic relatedness as a predictor of social interaction rates has received little attention as a potential driver of ASF transmission. A recent model of ASF transmission in wild boar highlighted a significant role of social structure in shaping spatial and temporal dynamics of ASF spread and showed that most transmission events occurred within family groups and within close distance of less than 1.5 km (Pepin et al., 2021). However, real-time infection and contact tracing data that could validate predictions from models of surveillance data are notoriously difficult to obtain from field studies and no such data exist for the wild boar—ASF system. Here, we used genetic relatedness as a proxy of social interactions as those two have been shown to correlate in the kin-based wild boar society (Podgórski, Lusseau et al., 2014). Kinship has been shown to predict infection risk in other wildlife disease systems, for example, chronic wasting disease in white-tailed deer (Grear et al., 2010) or bovine tuberculosis in badgers (Benton et al., 2016).
Previous studies have shown that the probability of ASF occurrence in wild boar populations increases with proximity to previous cases at a coarse spatial scale (>10 km) (Podgórski et al., 2020), while transmission rates appear to be highest at fine scales (<2 km (Pepin et al., 2021). Here, we investigate whether distance-dependent infection risk is influenced by genetic relatedness at a local spatial scale where relatedness might influence contact structure and, thus, impact disease transmission. We hypothesized that the infection risk would correlate positively with proximity and relatedness to ASF-positive individuals. We expected relationships of infection risk and proximity or genetic relatedness to become weaker with increasing the distance between individuals due to decay in contact rates (Pepin et al., 2016; Podgórski et al., 2018) and genetic similarity (Podgórski, Scandura et al., 2014; Poteaux et al., 2009). Additionally, we explored the differences in effects of relatedness and spatial proximity on infection probability between direct and indirect transmission. We expected that infection risk in both transmission modes will be distance-dependent but that relatedness would have a greater effect on infection risk among live animals compared to carcass-based transmission.
2 MATERIALS AND METHODS
2.1 Study area and sample collection
The study was conducted in north-eastern Poland where ASF was introduced in February 2014, <1 km from the border with Belarus, and has subsequently spread through the region in wild boar (Podgórski et al., 2020) and domestic pigs (Taylor et al., 2021). By mid-2016, ASF cases in wild boar were distributed continuously in the infected region covering about 4500 km2 and this is where samples used in this study originate (Figure 1). The area is mostly field-woodland mosaic characterized by extensive agriculture and low human population density (60 people/km2) and relatively high forest cover (31%). The landscape is flat (highest elevation 298 ms a.s.l.) with no significant natural or anthropogenic barriers to wild boar movement. Wild boar are distributed continually throughout the area with densities at the start of the ASF epidemic ranging between 0.5 and 5 ind./km2 at the forest district level (Regional Directorate of State Forests, Białystok, Poland).
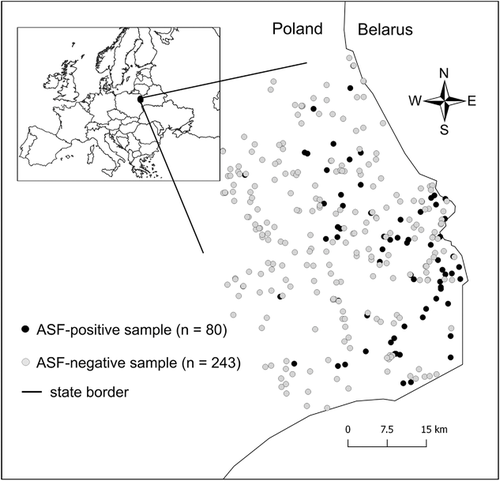
Following the introduction of ASF, an intensive surveillance program was implemented in the affected area. The program conducted laboratory tests of all hunted wild boar (active surveillance) and all wild boar found dead (passive surveillance). We used surveillance data routinely collected by the National Reference Laboratory for ASF at the National Veterinary Research Institute in Puławy, Poland. Wild boar samples were classified as ASF-positive (hereafter ‘case’) if presence of viral DNA was confirmed in real-time PCR or antibodies were detected using an ELISA test and confirmed with a immunoperoxidase test. Detailed description of the surveillance design and laboratory procedures can be found in Woźniakowski et al. (2016). A total of 5487 wild boars were sampled in the study area from February 2014 to July 2016 and 168 of them tested positive for ASF. It was not technically possible to genotype all of the sampled individuals. Instead, we selected a subsample of 1078 individuals (including all ASF-positive) evenly distributed across the study area for further analyses. Even distribution of samples was secured by selecting ASF-negative samples in a 10 km buffer around each ASF case which were distributed continuously in the study area, that is, without separate spatial clusters located beyond the capacity of wild boar movement. However, many samples, particularly originating from the carcasses, were of poor quality and yielded too few microsatellite loci (those yielding less than 13 out of 16 loci were excluded). Finally, we were able to satisfactorily genotype 323 samples (243 ASF-negative and 80 ASF-positive) which comprised a final dataset.
2.2 Microsatellite genotyping
DNA was extracted from tissue and blood using the Sherlock AX Kit (A&A Biotechnology, Gdynia, Poland), following the manufacturer's protocol. The extracts were quantified with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). All individuals were genotyped with a panel of 16 polymorphic microsatellite loci (S090, SW72, S155, S026, S355, S215, SW951, SW857, SW24, SW122, IGF1, SW461, SW1492, SW2021, SW2496, SW2532), which had been successfully used to study relatedness and genetic variation in wild boar populations from the study area (Podgórski, Lusseau et al., 2014; Podgórski, Scandura et al., 2014). The 16 autosomal STR loci were amplified in two independent multiplexed mixes using Qiagen Master Mix (Qiagen Inc, Hilden, Germany) reagents and fluorescently label primers. For the first panel, the fluorochromes were used: 6-FAM for the loci SW72, SW857; VIC for SW1492, IGF1; NED for S026, S215; PET for SW2021, SW2532 and for second panel 6-FAM for SW122, S0355; VIC for SW461, SW2496; NED for W24, S0155 and PET for SW951 and S0090. PCR was performed on Veriti® Thermal Cycler amplifier (Applied Biosystems, Foster City, CA, USA) using the following thermal profile: 5 min. of initial DNA denaturation at 95°C, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 90 s, elongation of starters at 72°C for 30 s and final elongation of starters at 60°C for 30 min. Analysis of the obtained PCR products was performed using an ABI 3130xl capillary sequencer (Applied Biosystems) at the Institute of National Research Institute of Animal Production (Cracow, Poland). The amplified DNA fragments were subjected to electrophoresis in 7% denaturing POP-7 polyacrylamide gel in the presence of a standard length of 500 Liz and a reference sample. The results of the electrophoretic separation were analyzed automatically using the GeneMapper ® Software 4.0 (Applied Biosystems).
2.3 Analysis of relatedness and spatial-genetic structure
Basic parameters of microsatellite polymorphism and genetic diversity were calculated using GENALEX 6.5 (Peakall & Smouse, 2006) and FSTAT (Goudet, 1995). GENEPOP 4.7 (Raymond & Rousset, 1995) was used to test loci for departures from linkage equilibrium and Hardy–Weinberg equilibrium (HWE) using the Markov chain method (parameters: 1000 dememorization steps, 100 batches, 1000 iterations per batch). The significance level was adjusted for multiple testing across loci using the sequential Bonferroni's correction (Rice, 1989).
Pairwise genetic relatedness (Queller & Goodnight, 1989) was calculated with GENALEX 6.5 among all individuals (n = 323) and subsequently used in all analyses. The Queller and Goodnight estimator is an unbiased measure of relatedness based on population allele frequencies. It ranges from −1 to 1, where positive values indicate pairs more related than average and negative less related than average. We used the Queller and Goodnight estimator to allow comparison with previous studies of wild boar social structure which we build upon (Podgórski, Scandura et al., 2014). We used dyadic relatedness as continuous predictor in models explaining variation in infection probability. To facilitate interpretation of the results, we adopted three biologically meaningful levels of relatedness: 0 (unrelated, average inter-group relatedness), 0.25 (2nd degree relatives, average intra-group relatedness), 0.5 (1st degree relatives, full families) based on previously published relatedness data (Podgórski, Scandura et al., 2014) and dychotomized highly related (hereafter ‘kin’) and un-related (hereafter ‘non-kin’) pairs of individuals using mean within-group relatedness of 0.247 (Podgórski, Scandura et al., 2014). Analysis of spatial-genetic structure was performed with GENALEX 6.5. Autocorrelation coefficients (r) between pairwise genetic and geographic distance matrices were calculated for pre-defined Euclidean distance classes (Supporting Information Figure S1) which correspond to spatial-genetic structure previously observed in wild boar populations (Pepin et al., 2016; Podgórski et al., 2018; Podgórski, Scandura et al., 2014; Poteaux et al., 2009). Spatial-genetic structure was examined among all individuals as well as among ASF-positive and ASF-negative individuals separately. Spatial autocorrelation coefficients were compared across distance classes using randomization tests (10,000 permutations) (Manly, 1997).
2.4 Estimating probability of infection
For every sampled individual (n = 323), we selected all other individuals within a pre-defined temporal window of potential transmission set at 30 days prior to sampling. This period reflects the upper limit of lifespan after ASF infection, that is, infection-to-death time (Gallardo et al., 2017; Pietschmann et al., 2015). For samples originating from carcasses, the window was set to 60 days prior to sampling and included infection-to-death time and carcass decomposition time during which the sample was taken. A decomposition time of 30 days was chosen arbitrarily because the decomposition status of most carcasses was unknown and the exact date of death could not be determined. This time frame was conservative given usually longer decomposition times of wild boar tissues (Probst et al., 2020) and ASF virus persistence in them (Fischer et al., 2020). Additionally, the temporal window was extended by 7 days post-sampling of the focal individual to account for the retrieval and reporting lag of the ASF-positives. This selection resulted in 7111 pairs of individuals who were used in further analysis.
First, we compared relatedness of individuals with paired infections (pairs where both individuals were infected) and without paired infection (none or one individual of the pair were infected) within four distance classes (0–2, 2–5, 5–10, 10–20 km). Relatedness levels were compared using randomization tests (10,000 permutations) (Manly, 1997). Four distance classes were selected based on current knowledge of wild boar socio-spatial ecology: (1) ‘high-contact’ zone (0–2 km): social contacts among individuals are most frequent, both within and between groups (Podgórski et al., 2018; Yang et al., 2020), (2) ‘medium-contact’ zone (2–5 km): interactions among neighbouring social groups (Pepin et al., 2016; Podgórski et al., 2018), (3) ‘low-contact’ zone (5–10 km): sporadic contacts between distant groups with non-overlapping home ranges, distance of most natal dispersal (Keuling et al., 2010; Podgórski, Scandura et al., 2014; Prévot & Licoppe, 2013), (4) ‘no-contact’ zone (>10 km): groups do not interact, occasional long-distance movements (Andrzejewski & Jezierski, 1978; Podgórski, Scandura et al., 2014). Those distance classes were used to help interpret modelling results.
Second, we selected pairs of the focal individual (ASF-positive or ASF-negative) and ASF-positive individual (hereafter carrier) (n = 1928 pairs). However, for further modelling we only retained pairs of individuals who were 20 km or less apart (n = 705) because this spatial scale was relevant for our questions and allowed investigating how genetic relatedness and contact heterogeneity relate to disease transmission at a local scale. The geographic distance (Supporting information Figure S2) and genetic relatedness (Supporting information Figure S3) between individuals in each pair were used as covariates explaining the binary response of ASF infection status of the focal individual (0—negative, 1—positive). These two covariates were weakly correlated (Spearman's R = −0.11).
We analyzed the effects of distance and relatedness on the probability of ASF infection using a generalized additive model with a binomial error structure (GAM 1) in the ‘mgcv’ package implemented in R (Wood, 2020). We set ASF infection status of the focal individual as a response variable. Explanatory variables included both main and interactive effects of geographic distance and genetic relatedness between the focal individual and ASF carrier that were fitted as non-parametric smoothing term. We assumed the same level of curvilinearity in the effect of geographic distance and genetic relatedness, thus, we set the same smoothing range. The probability of ASF infection reflects the chance of any sampled individual from the study area testing positive for ASF within the 2.5 years of the study period based on the covariates. Next, we investigated whether the origin of the ASF carrier (dead or alive), that is, the transmission mode, influenced the relationship between proximity or relatedness and infection risk. To identify if transmission mode shaped the effect of distance and relatedness on the probability of ASF infection, we fitted GAM 2 with the same error structure and smoothing settings as in GAM 1. To get separate results for the two transmission modes, apart from the variables used in GAM 1, we added transmission mode (0–sample collected from live, i.e., hunted) individual and 1–sample collected from dead individual, i.e., carcass) as an additional variable implemented to the model formulation with argument ‘by’. All statistical analyses were performed in R 4.0.2 (R Core Team, 2020).
3 RESULTS
3.1 Spatial-genetic structure and relatedness
In total, 135 alleles were detected across 16 analyzed loci. All loci were polymorphic with the number of alleles per locus ranging from 3 to 18 (mean ± SE: 8.4 ± 0.94). Missing data (i.e., % of missing alleles) amounted to 2.4% of the dataset and no individual was typed at less than 13 loci. Expected and observed heterozygosity averaged 0.61 ± 0.06 and 0.56 ± 0.06, respectively. Following sequential Bonferroni's corrections, the overall population showed deviation from HWE at eight loci and from linkage equilibrium in 2 out of 120 pairs of loci. Such deviation from equilibrium was most likely attributed to the inherent substructure of the population (i.e., presence of kin groups) and all loci were retained for statistical analyses.
Spatial autocorrelation analysis revealed the presence of genetic structure, as indicated by an autocorrelation coefficient (r)greater than at random, at a distance of up to 20 km at the population level and up to 10 km among ASF-positive individuals, with genetic similarity between individuals declining from the smallest distance (Supporting information Figure S1). Genetic structuring was stronger among ASF-positive individuals compared to ASF-negative ones within each distance class up to 10 km (0–2.5 km distance class: rpos = 0.078 ± 0.003 and rneg = 0.033 ± 0.001, p < .001; 2.5–5 km distance class: rpos = 0.028 ± 0.002 and rneg = 0.015 ± 0.001, p < .001; 5–10 km distance class: rpos = 0.019 ± 0.001 and rneg = 0.011 ± 0.0004, p < .001; Supporting information Figure S1).
Considering the entire study area, the coefficient of relatedness in the population averaged 0.0003 ± 0.002 and relatedness among individuals with paired infections was much higher than among those without paired infections (0.049 ± 0.009 and −0.003 ± 0.002, respectively, p < .001). The differences were influenced by the distance between pairs (Figure 2). Relatedness among animals with paired infections was highest in the 0–2 km and 2–5 km distance classes (0.129 ± 0.054 and 0.129 ± 0.055, respectively) and lower in the subsequent classes (5–10 km: 0.018 ± 0.017;10–20 km: 0.030 ± 0.014). Relatedness between individuals with paired infections was consistently higher than among those without paired infections, except within the 5–10 km distance class (Figure 2).
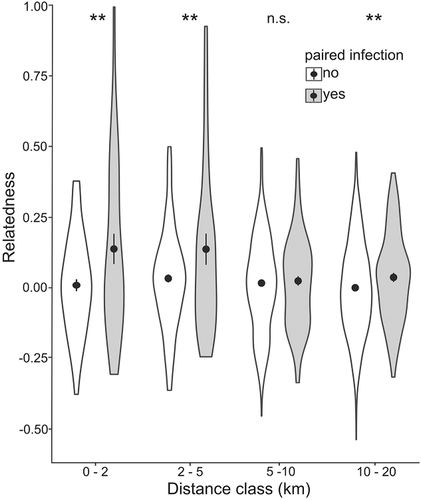
3.2 Probability of infection
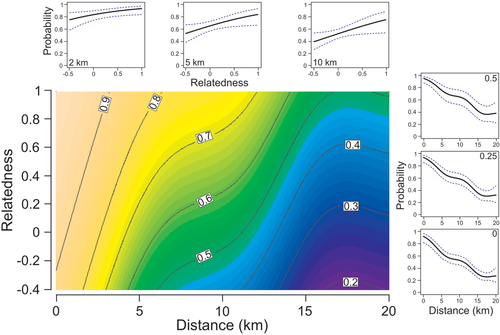
We found a positive relationship between ASF infection probability and proximity (X2 = 31.33, p < .001) as well as relatedness (X2 = 4.71, p = .03) to infected individuals (Table 1; Figure 3). At short distances to ASF carriers (0–2 km, high-contact zone), mean (± SE) probability of infection was 90 ± 0.3%. At further distances of 2–5 km (medium-contact zone) infection risk averaged 78 ± 0.7%, while within distances of 5–10 km (low-contact zone) and 10–20 km (no-contact zone) mean probability of infection decreased to 64 ± 0.9% and 42 ± 1%, respectively. Effect of relatedness was not consistent across the range of inter-individual distances, as indicated by significant interaction term (X2 = 1.16, p = .029; Table 1). At the distance of 2 km, infection risk varied only slightly across relatedness to ASF carriers; ranging from 84% at relatedness 0 (unrelated individuals, from different groups), to 87% at relatedness 0.25 (2nd degree relatives, group members), to 89% at relatedness 0.5 (1st degree relatives, full families) (Figure 3). This indicates that within 2 km variation in infection risk was shaped by the presence of infected individuals rather than by relatedness to them. However, infection risk among kin (relatedness ≥ 0.25) within 2 km was high. All of the animals which were surrounded by ASF-positive kin (n = 17) tested positive, while none of the animals surrounded by ASF-negative kin (n = 7) tested positive. Among animals surrounded only by infected non-kin between 0 and 2 km (n = 18), the proportion of ASF-positive individuals was 83%, indicating that at distances within 2 km individuals were 17% less likely to become infected by a non-kin individual compared with kin individual on average. At the distance of 5 km, infection risk ranged from 65% at relatedness 0 to an ASF carrier, to 70% at relatedness 0.25, to 76% at relatedness 0.5 (Figure 3). At the distance of 10 km, infection risk ranged from 53% at relatedness 0, to 59% at relatedness 0.25, to 65% at relatedness 0.5 (Figure 3). The effect of relatedness to ASF carriers tended to be more pronounced at larger distances. The difference in infection risk from kin and non-kin carriers was 6% on average (93 and 87%, respectively) in the high-contact zone (0–2 km), 12% (84 and 72%) in the medium-contact zone (2–5 km), 16% (71 and 55%) in the low-contact zone (5–10 km) and 17% (50 and 33%) in the no-contact zone (10–20 km).
| Variable | Estimate ± SE or edf | Test statistics | p-value | R2adj | AIC |
|---|---|---|---|---|---|
| GAM 1 | 0.15 | 865.0 | |||
| Parametric terms | z | ||||
| Intercept | −0.005 ± 0.09 | −0.06 | .95 | ||
| Smooth terms | Χ2 | ||||
| Distance | 2.86 | 31.3 | <.001 | ||
| Relatedness | 1.00 | 4.71 | .03 | ||
| Distance × Relatedness | 1.06 | 1.16 | .03 | ||
| GAM 2 | 0.16 | 857.9 | |||
| Parametric term | z | ||||
| Intercept | 0.12 ± 0.09 | 1.31 | .19 | ||
| Sample type (carcass vs. hunted) | −0.82 ± 0.25 | −3.29 | .001 | ||
| Smooth terms | Χ2 | ||||
| Distance | 2.95 | 5.81 | .18 | ||
| Relatedness | 0.0001 | 0 | .99 | ||
| Distance × Relatedness (carcass) | 2.00 | 7.26 | .03 | ||
| Distance × Relatedness (hunted) | 2.51 | 7.23 | .08 | ||
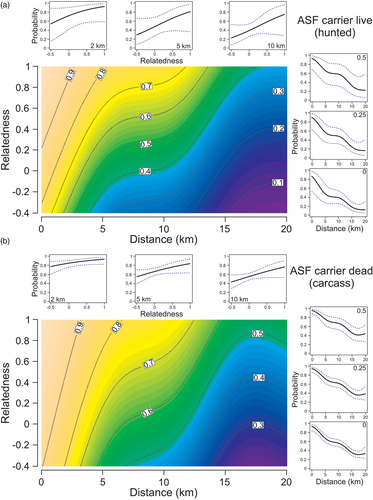
We found that relatedness and proximity affected infection risk differently depending on whether the carrier was sampled dead (carcass) or alive (hunted). The importance of transmission mode was confirmed by the significantly better fit and lower AIC values of the model split by carrier sample type (GAM 2) compared with less complex main model (GAM 1) (ANOVA: Χ2 = 14.7, p = .006; Table 1). Effect of relatedness on infection risk was less consistent across the range of inter-individual distances for carcass-based transmission, as indicated by a significant interaction term (X2 = 7.26, p = .027; Table 1), in comparison to direct transmission (i.e., from hunted carriers), as indicated by a marginally significant interaction term (X2 = 7.23, p = .08; Table 1). Relatedness tended to have a larger effect on the probability of infections acquired through direct transmission than through carcass-based transmission, particularly at close distances (Figure 4). For paired infections between live individuals, the difference in infection risk from kin and non-kin carriers was 13% on average at 0–2 km (90 and 77%, respectively), 23% at 2–5 km (77 and 54%) and 26% at 5–10 km (64 and 38%). For paired infections between a carcass and a live individual, the difference in infection risk from kin and non-kin carriers was 5% at 0–2 km (93 and 88%, respectively), 11% at 2–5 km (84 and 73%) and 15% at 5–10 km (72 and 57%).
4 DISCUSSION
Our results showed that the risk of ASF infection was shaped by interacting effects of spatial proximity and genetic relatedness to infected individuals. While both proximity and relatedness to ASF carriers were positively associated with infection risk, the effect of proximity was generally stronger. Relatedness tended to have a weaker effect at close distances. In the high-contact zone (0–2 km), where frequency of social interactions and probability of encountering a carcass are high, infection risk was shaped by the presence of infected individuals rather than by relatedness to them. Nevertheless, infections were more frequent among close relatives, that is, kin or group members, indicating that familial relationships could have played a significant role in ASF transmission. In the medium-contact zone (2–5 km), infection risk decreased but was still associated with relatedness and paired infections were more frequent among more related individuals. At larger distances, infection risk further declined with relatedness and proximity to carriers, dropping by 60% from relatives in the high-contact zone (0–2 km) to un-related individuals in the no-contact zone (10–20 km). Transmission mode influenced the relationship between proximity or relatedness and infection risk, which provides a novel insight into how transmission mechanisms determine variation in ASF incidence in wild boar. While distance played a major role in predicting infection risk in both transmission modes, relatedness tended to have a larger effect on the probability of infections acquired from live carriers than from infected carcasses, particularly at close distances to ASF carrier.
Social contacts directed toward relatives can lead to local clustering of disease prevalence (Blanchong et al., 2007; Delahay et al., 2000). Social relationships in wild boar tend to correlate with genetic relatedness and are the strongest among closely related group members (Gabor et al., 1999; Podgórski et al., 2018; Podgórski, Scandura et al., 2014). Therefore, we predicted a strong effect of relatedness on disease transmission at close distances (0–2 km) due to socially-driven contact heterogeneity. Indeed, ASF infection risk was the highest at close distances but the effect of relatedness was weaker than at further distances. This suggests a dominant role of proximity to ASF carriers over relatedness to them. However, our descriptive analysis found that individuals who were infected simultaneously (i.e., paired infections) tended to be more related than those un-infected. This trend was particularly noticeable at the upper range of relatedness distribution, that is, among close kin or group members. This apparent inconsistency could be explained by a smaller number of positive samples from hunted individuals relative to those found dead (16 and 84%, respectively). Our results showed that relatedness tended to have a larger effect on the probability of infections acquired from live carriers, particularly, at close distances. Such a pattern is consistent with kin-biased associations in wild boar manifested in more regular and longer lasting contacts with relatives (Podgórski, Lusseau et al., 2014; Poteaux et al., 2009) which can facilitate disease transmission. On the other hand, inter- and intra-group contacts in wild boar occur most frequently at a similar spatial scale of 0–2 km (Podgórski et al., 2018; Yang et al., 2020). This spatial overlap in social connectivity can lead to the highest transmission rates in the high-contact zone, as observed in this study and previously (Pepin et al., 2021) while confounding the effects of inter- versus intra-group transmission (i.e., relatedness). Indirect transmission through infected carcasses plays an important role in the spread and persistence of ASF virus (Chenais et al., 2018). This mechanism accounts for more than half of ASF infections in wild boar population (Pepin et al., 2020). In our study, infection risk from carcass was mostly distance-dependent and the effect of relatedness was relatively weak, especially in close proximity. Because diseased animals tend to die locally (within their home range), they will be a source of infection mainly for the most proximate individuals from their own or neighbouring social groups. Susceptible animals can come into contact with nearby infected carcass of the individual which was a member of a different but spatially overlapping social group and was not closely related. Abundance of infectious carcasses in the surrounding environment could, thus, lead to infection regardless of relatedness.
Positive association between infection risk and relatedness extended beyond the closest socio-spatial environment (>2 km) suggesting that kin relationships can coincide with higher transmission rates even if inter-individual contacts are less frequent. This pattern is unlikely to have resulted from the dispersing individuals infected in the natal groups because it typically takes longer to disperse (Podgórski, Scandura et al., 2014) than it takes the disease to hamper movements (Blome et al., 2013). To alleviate symptoms of the disease, infected wild boar seek specific habitats which differ from those regularly used (Morelle et al., 2019). These preferences and restricted mobility of sick animals can separate them from the group and result in dispersion of diseased group members and wide distribution of samples from related individuals. Additionally, kin-directed interactions over larger distances could be maintained by temporal fission-fusion events of core groups, similar to that observed in African elephants (Archie et al., 2006) or giraffes (Carter et al., 2013). These dynamics could provide a mechanism for disease transmission among distant relatives. However, fission-fusion dynamics in wild boar has not been systematically studied and it is difficult to tell whether temporal scales of social and disease dynamics would match and help explain patterns observed in our study. Direct transmission is unlikely to play a significant role in shaping infection risk at distances over 5 km since inter-group contacts are very rare at those distances (Pepin et al., 2016; Podgórski et al., 2018). While a distance of 5–10 km exceeds the size of typical home range, it is within a range of daily travel (Podgórski et al., 2013) and could be covered during dispersal, foraging or mating forays leading to distant transmission events. These behaviours, however, are not typically seeking contact with kin and particularly in case of mating or dispersal are often seeking non-kin to avoid inbreeding (Archie et al., 2007; Biosa et al., 2015; Hoffman et al., 2007). The observed effect of distant infected individuals on infection risk could be also a by-product of correlated local enzootic dynamics (i.e., spatial and temporal co-occurrence of cases) rather than direct transmission. Spatial clusters of increased ASF prevalence identified previously were measured at 20–40 km (Podgórski et al., 2020; Taylor et al., 2021). However, these studies used data aggregated over time periods (months-years) exceeding temporal windows of ASF transmission (days-weeks) and, thus, could not capture fine-scale disease dynamics well. Our results indicate that ASF outbreaks in wild boar are highly localized. This is supported by strong genetic structuring among infected animals (Supporting information Figure S1) and rapid decay of infection probability with distance (71% at 5 km, 60% at 10 km, 34 % at 20 km; Figure 3). The spatial limits of transmission highlight the possibility to control outbreaks if containment measures, such as fencing, zoning and carcass removal, are employed immediately and early detection is ensured by effective surveillance. Our results show that ASF infection risk on the landscape was mostly distance-dependent with a modulating effect of relatedness. Therefore, it appears more practical and cost-efficient to apply distance-based measures for disease control than population genetics tools.
Together our results show that ASF infection risk declines with distance, matching spatial changes in contact intensity and proximity to infectious carcasses. Infection-causing contacts correlated with relatedness particularly among live animals. At close distances, infections were more frequent among close kin while at medium distances relatedness predicted infection risk more consistently. This indicates that physical kin relationships can extend beyond the immediate social environment and induce differential transmission rates, similarly to transmission of chronic wasting disease in white-tailed deer (Grear et al., 2010). Nevertheless, infection risk was primarily influenced by the distance to infected individuals and carcasses, conforming to the previous modelling study which found that most transmission events occur within <1.5 km with some rare events at longer distances (Pepin et al., 2021). In future studies, it may be informative to examine the potential importance of other variables, such as seasonality in social dynamics, movements and demography to explain variation in infection probability that is unexplained by our models.
ACKNOWLEDGEMENTS
This work was supported by the National Science Centre, Poland (Grant number 2014/15/B/NZ9/01933) and Ministry of Agriculture, Czech Republic (Grant number QK1910462). We thank M. Niedziałkowska from the Mammal Research Institute, Polish Academy of Sciences, for help in genetic analyses.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
Samples used in this study were collected within the national ASF surveillance programme and follows relevant legislation regarding animal treatment.
Open Research
DATA AVAILABILITY STATEMENT
Data available on request from the authors.




