Pathogenic investigations of Streptococcus pasteurianus, an underreported zoonotic pathogen, isolated from a diseased piglet with meningitis
Abstract
Streptococcus pasteurianus, an underreported opportunistic pathogen, is considered an increasingly recognized cause of meningitis and bacteremia in many animals and humans worldwide. However, except for some epidemiological studies, there is no report about the gene-deletion mutagenesis, virulence factors, reservoir niches or animal infection models for this pathogen. In this study, we first isolated an S. pasteurianus strain from a newly weaned piglet's brain with meningitis. The genomic sequence of this swine isolate WUSP067 shared high homology with that of two human strains. The comparative genome analysis showed that strain WUSP067 contained a fucose utilization cluster absent in human strains, and it shared 91% identity with that of an integrative and conjugative element (ICE) ICEssuZJ20091101-2 from Streptococcus suis, another important swine bacterial pathogen. Strain WUSP067 was resistant to erythromycin, tulathromycin, lincomycin, clindamycin, doxycycline and gentamycin, and ICEs are vehicles for harbouring antimicrobial resistance genes. The infection model was established using the 3-week-old newly weaned ICR mice. The 50% lethal dose value of strain WUSP067 was 4.0 × 107 colony-forming units per mouse. The infected mice showed severe signs of meningitis and pathological changes in brains. Furthermore, the capsule-deficient mutant was generated using natural transformation, and we showed that capsule was an essential virulence factor for S. pasteurianus. In addition, we found that tonsils and hilar lymph nodes of healthy pigs may be reservoir niches for this bacterium. Thus, our study provided valuable information about the pathogenetic characteristics and antimicrobial resistance of S. pasteurianus and paved the way for studying its pathogenesis.
1 INTRODUCTION
Streptococcus pasteurianus, previously known as Streptococcus bovis biotype II/2, is an opportunistic pathogen in many animals, causing meningitis in ducklings and calves and septicemia in goslings and turkey poults, leading to high mortality (Barnett et al., 2008; Li et al., 2013; Saumya et al., 2014; Trotta et al., 2019). In addition, this bacterium is also recognized as an emerging human pathogen (Gherardi et al., 2016), causing uterine infection, endometritis, meningitis, bacteremia, septicemia or even death, mainly in immunodeficient patients, older people, neonates and pregnant women (Binghuai et al., 2013; Gherardi et al., 2016; Hede et al., 2015; Klatte et al., 2012; Matesanz et al., 2015; Sturt et al., 2010). S. pasteurianus may also be associated with colonic malignancies (Chand et al., 2016; Takamura et al., 2014). This bacterium was proposed as S. pasteurianus (Poyart et al., 2002) or Streptococcus gallolyticus subsp. pasteurianus (Schlegel et al., 2003). Although the discrepancy exists whether S. pasteurianus is a species or subspecies, both classifications are currently recommended (Jans et al., 2015). Since the DNA–DNA hybridization values between S. pasteurianus and S. gallolyticus (65% and 57%) are significantly below the usual standard of >70% to denote these as the same species (Schlegel et al., 2003), the classification of S. pasteurianus was used in this study. Except for the above epidemiological studies, very little is known for this bacterium. So far, the genome of two human isolates ATCC 43144 and NCTC 13784 has been sequenced. However, there is no report about the gene-deletion mutagenesis, virulence factors, reservoir niches or animal infection models for this important zoonotic pathogen, which dramatically hinders understanding its pathogenesis.
In this study, we first isolated an S. pasteurianus strain from a newly weaned piglet's brain with meningitis. The antimicrobial resistance and pathogenicity characteristics of S. pasteurianus strain WUSP067 were investigated, including the genomic analysis, gene manipulation technique, infection model, reservoir niche, virulence factor and transmission patterns of antibiotic resistance genes.
2 MATERIALS AND METHODS
2.1 Bacterial isolation and culture conditions
S. pasteurianus WUSP067 was isolated from the brain of a newly weaned piglet with meningitis. S. pasteurianus was cultured in brain heart infusion (BHI, Becton, Dickinson and Company) liquid or agar media at 37°C with 5% CO2. The isolation of S. pasteurianus from a piglet's brain was also carried out on a BHI agar plate. When screening or culturing the deletion mutant of S. pasteurianus, appropriate spectinomycin (Spc, 100 μg/ml Sigma) was added. For studying fucose or glucose utilization, bacteria were grown in BHI to mid-exponential phase (OD600 = 0.6), washed twice in PBS, and then adjusted to OD600 of 0.6 with Dulbecco's Modified Eagle Medium without glucose (DMEM, Solarbio) containing fucose (Macklin) with a final concentration of 2%, or glucose (Kermel) with a final concentration of 1%. The growth of S. pasteurianus without O2 was carried out in a 1.5-ml microtube containing 1.4 ml of culture sealed by parafilm and shaken at 180 rpm. The growth of S. pasteurianus with O2 was carried out in a 50-ml flask containing 10 ml of culture shaken at 180 rpm.
2.2 Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of macrolides [erythromycin (Macklin, 98% purity) and tulathromycin (TMstandard, 96.2% purity)], aminoglycosides (gentamycin) (Macklin, USP level), lincosamides [clindamycin (Macklin, 99% purity) and lincomycin (Macklin, 98% purity)] and tetracyclines (doxycycline) (Macklin, 99% purity) to S. pasteurianus was determined using the broth microdilution method following Clinical Laboratory Standards Institute document (CLSI, 2020). The breakpoint for resistance to erythromycin and clindamycin was based on the CLSI for Streptococcus viridans group (Li et al., 2019). Since the breakpoint for resistance to the rest of antimicrobial agents is not available, the breakpoint for resistance to the rest of antimicrobial agents was adopted according to the previous study (Wang et al., 2021).
2.3 Genome sequencing, assembly, annotation and bioinformatics analysis
According to the manufacturer's protocol, the total genomic DNA of strain WUSP067 was extracted using a Bacterial DNA Kit (OMEGA). The genome of strain WUSP067 was sequenced by Illumina NovaSeq PE150 at Novogene Bioinformatics Technology Co., Ltd. The software, including SPAdes, SOAPdenovo (version 2.04), Gapclose, ABySS and CISA (v1.12), assembled the genome according to the company's protocol. The genome was annotated through the NCBI prokaryotic annotation pipeline. The sequence and annotation of strain WUSP067 have been deposited in NCBI (Accession No. NZ_CP039457.1). The average nucleotide identity (ANI) was calculated by OrthoANI (Lee et al., 2015). Island Viewer 4 was used to predict possible GIs (http://www.pathogenomics.sfu.ca/islandviewer/). Antibiotic resistance genes were analysed by ResFinder 4.0 (Bortolaia et al., 2020). The prophage was predicted using PHAST (Zhou et al., 2011). The ICE structure was predicted using ICEberg 2.0 (Liu et al., 2019). Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas and restriction-modification (RM) systems were analysed using CRISPRCas Finder (https://crisprcas.i2bc.paris-saclay.fr/) and REBASE (http://rebase.neb.com/rebase/rebase.html), respectively.
2.4 Identification of S. pasteurianus and Streptococcus suis in healthy pig tonsils and hilar lymph nodes
The tonsils and hilar lymph nodes from healthy pigs were collected from slaughterhouses in Chongqing and Xishuangbanna cities, respectively. The samples were washed with sterile PBS. Three pieces (0.1 g per piece) were taken from each sample, mixed in sterile PBS with the ratio of 1:9 (wt/vol), and then processed in a Homogenizer (MP biomedicals). The homogenate was transferred to BHI with 15 mg/L polymyxin and 30 mg/L nalidixic acid at a ratio of 1:50 and incubated at 37°C with 5% CO2 for 8 h. Bacteria were centrifuged at 8000 × g for 5 min. The genomic DNA of bacteria was extracted using a TIANamp Bacteria DNA Kit (TIANGEN) according to the manufacturer's instructions. The gene SGPB0680 (E8M05_RS04035 in strain WUSP067) was used to identify S. pasteurianus in the previous study (Hatrongjit et al., 2017). In the present study, in addition to gene SGPB0680, two S. pasteurianus specific genes (E8M05_RS05155 and E8M05_RS06300 in strain WUSP067) were also included. Samples with the presence of all three genes were judged to be positive for S. pasteurianus. The primers are listed in Table 1. To isolate S. pasteurianus, bacteria from S. pasteurianus positive samples were streaked on BHI agar plates with 15 mg/L polymyxin and 30 mg/L nalidixic acid. Thirty colonies from each sample were selected to identify S. pasteurianus by a PCR assay to detect the above three specific genes.
| Genea | Primer sequence (5′−3′) | Size (bp) | Description |
|---|---|---|---|
| E8M05_RS04035 | GGTGTGCCAGATGGACAAGA | 531 | Cell wall surface protein |
| CGTTACCGTTGTTCCGCTTG | |||
| E8M05_RS05155 | CGCAAATCTTTTCTTTATAACAGG | 714 | MFS transporter |
| TTAAAAGCACCGATTCTATCCAT | |||
| E8M05_RS06300 | TCTTCAACCACAAGCGGTGA | 607 | Carboxylesterase /lipase family protein |
| CTACAGCAAATCGATGGCAAG |
- a The genes are from S. pasteurianus strain WUSP067.
For identifying S. suis, the homogenate was transferred to Todd–Hewitt broth with 1.2 mg/L crystal violet, 30 mg/L nalidixic acid and 1.2 mg/L gentamicin at a ratio of 1:50 and incubated at 37°C with 5% CO2 for 8 h. The genes recN encoding recombination/repair protein and gdh encoding glutamate dehydrogenase were used to identify S. suis. The samples with the presence of both recN and gdh are considered as S. suis-positive samples.
2.5 Construction of cpsE deletion mutant (ΔcpsE)
The deletion mutant ΔcpsE was achieved by the method of natural transformation. The upstream and downstream fragments of the deletion region were amplified by PCR with primers ΔcpsE-A/B and ΔcpsE-C/D from the genomic DNA of strain WUSP067, respectively. The spectinomycin-resistant gene spc was amplified by primers spc-F/R from the plasmid pSET4s. The above three amplification products were then ligated by fusion PCR with primers ΔcpsE-A/D. The synthesized ComS7-15 peptide of strain WUSP067 (IVLTGWWGV, 250 μM) (ChinaPeptides Co., Ltd.) and the fusion fragment (1.0 μg) were added to 100 μl strain WUSP067 (OD600 = 0.02-0.07). The mixture was incubated at 37°C without shaking for 2 h and then plated in BHI agar medium containing spectinomycin. The deletion mutant was detected by PCR with primers ΔcpsE-G1/G2 and ΔcpsE-H1/H2 and was further confirmed by sequencing. The specific primers for PCR are listed in Supplementary Table S1.
2.6 Transmission electron microscopy (TEM)
The TEM analysis was performed to investigate if ΔcpsE losses the capsule according to a previous study (Li, Fu, et al., 2017). In brief, bacteria were harvested at the mid-exponential phase by centrifugation and fixed in 5% glutaraldehyde at room temperature for 2 h. According to the manufacturer's instruction, the capsule of S. pasteurianus was examined with a JEM-1010 TEM (JEOL, Ltd., Japan).
2.7 Mice infection experiments
Three-week-old newly weaned specific pathogen-free (SPF) ICR mice (SiPeiFu) were used for infection experiments. Bacteria were grown in BHI to mid-exponential phase (OD600 = 0.6), washed twice in PBS, and then adjusted to the appropriate doses. For determining the 50% lethal dose (LD50) value, four groups of mice (ten mice per group) were intraperitoneally injected with strain WUSP067 with doses of 1.5×109, 3×108, 6×107 or 1.2×107 colony-forming units (CFU) per mouse. Appropriate dilutions were plated on BHI agar plates to quantify the bacterial CFUs. Mortality was monitored once a day for 2 weeks post-infection. The LD50 value was calculated by the Reed and Muench method (Reed & Muench, 1938). Five dead mice were randomly selected for bacteriological investigations, and bacteria from the brain, heart, liver, spleen and lungs of these five dead mice were streaked on BHI agar plates by an inoculation loop. The plates were incubated at 37°C with 5% CO2. Five colonies from each plate were selected, and the isolation of S. pasteurianus was confirmed by a PCR assay to detect S. pasteurianus specific genes (E8M05_RS04035, E8M05_RS05155 and E8M05_RS06300). For determining the effect of strain WUSP067 on mice growth performance, two groups of mice (ten mice per group) were intraperitoneally injected with WUSP067 with doses of 6.4×107 or 1.7×106 CFU per mouse. The statistical analysis was performed with a two-tailed unpaired t-test. For histopathological examination, six mice were intraperitoneally injected with strain WUSP067 with doses of 1×107 or 1×108 CFU per mouse (three mice per dose). Mice in control groups were injected with PBS. The paraffin section was prepared according to our previous study (Zhang et al., 2018). The tissue was fixed with 4% paraformaldehyde for more than 24 h. The tissue was dehydrated by a series of alcohols, transparentized in xylene and embedded in paraffin. The tissue was then cut into 4 μm slices. The slice was stained with haematoxylin and eosin according to a previous study (Fischer et al., 2008). The stained slice was examined with an optical microscope (NIKON, ECLIPSE E100). For comparing the virulence of wild-type strain and ΔcpsE, two groups of mice (10 mice per group) were intraperitoneally injected with wild-type strain or ΔcpsE with a dose of 3×108 CFU per mouse. Mortality was monitored once a day for 2 weeks post-infection. Survival curves were analysed by the log-rank (Mantel–Cox) test.
2.8 Quantitative RT-PCR (RT-qPCR)
S. pasteurianus strain WUSP067 was cultured to the exponential growth phase (OD600 = 0.6) in BHI, washed twice in PBS and resuspended in the same volume of DMEM or DMEM containing fucose with a final concentration of 0.2%, 1% or 2% for 2 h. Bacteria were collected, and the total RNA was extracted using the FastRNA Pro Blue Kit (MP biomedicals) according to the manufacturer's protocol. The residual DNA was removed by DNase I (Takara). The RNA sample was stored at −80°C until needed.
The RT-qPCR analysis was performed according to our previous study (Wu et al., 2014). The cDNA was produced with a cDNA reverse transcription kit (Vazyme), and the RT-qPCR was conducted using the QuantStudio 6 Flex (Thermo Fisher Scientific) with the SYBR Premix Ex Taq™ (Takara) kit according to the manufacturer's protocol. The gene gyrA was used as the internal control. Three biological replicates were performed for each sample. The relative fold change was calculated based on the 2−ΔΔCT method. Experimental results were shown as mean ± standard error of the mean (SEM). The statistical analysis was performed with a two-tailed unpaired t-test. The specific primers for RT-qPCR are listed in Supplementary Table S1.
3 RESULTS
3.1 The genome sequence S. pasteurianus strain WUSP067 shares high homology with that of human isolates
-
The 16S rRNA sequence of strain WUSP067 shared 100% identity with that of S. pasteurianus type train NCTC 13784, a human isolate responsible for meningitis, and S. pasteurianus strain ATCC 43144 isolated from human blood.
-
The sodA gene encoding a superoxide dismutase was considered a more discriminative target sequence than the 16S rRNA gene for differentiating closely related species belonging to the Streptococcus genus (Poyart et al., 1998). The sodAint sequence (from positions 25 to 510) identity within S. pasteurianus cluster was ≥ 98.9% (Poyart et al., 2002). The sodAint sequence of strain WUSP067 shared 99.18% with that of type train NCTC 13784.
-
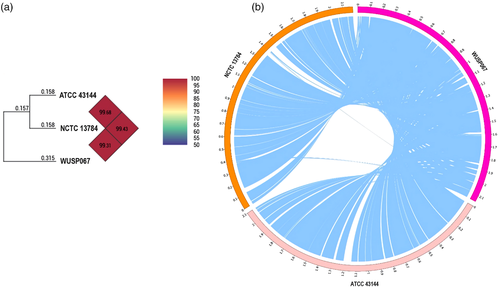
The 95−96% ANI threshold can be used as a boundary for bacterial species circumscription (Richter & Rossello-Mora, 2009). The ANI of strain WUSP067 shared 99.31% and 99.43% with that of human isolates NCTC 13784 and ATCC 43144, respectively (Figure 1a), confirming that strain WUSP067 belongs to S. pasteurianus.
-
Lin et al. (2011) listed S. pasteurianus specific gene candidates. These candidates were reanalysed by NCBI BLAST in this study, and 26 of them are only present in S. pasteurianus strains whose complete genomes are available in NCBI (Supplementary Table S3). Strain WUSP067 contains all these 26 genes. In addition to gene SGPB0680 (E8M05_RS04035 in strain WUSP067) (Hatrongjit et al., 2017), two other genes (E8M05_RS05155 and E8M05_RS06300) from these 26 genes were also used to identify S. pasteurianus in this study.
To determine the reservoir niches of S. pasteurianus and its carrier rate in healthy pigs, the tonsils and hilar lymph nodes from healthy pigs were tested. The detection rate of S. pasteurianus in pig tonsils and hilar lymph nodes was 12% (6/50) and 52% (26/50), respectively. The S. pasteurianus positive samples were further tested for the presence of S. suis. We found that three hilar lymph nodes were positive for these two bacteria. However, we cannot isolate S. pasteurianus from pig tonsils or hilar lymph nodes due to the vigorous growth of other bacteria and the relatively low abundance of S. pasteurianus in these samples.
3.2 Strain WUSP067 is multidrug resistant, and ICEs are vehicles for harbouring antimicrobial resistance genes
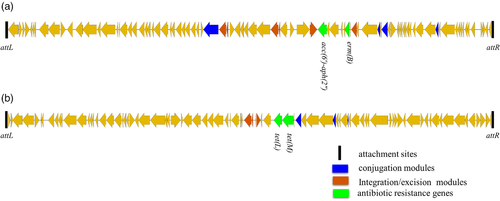
There are five antimicrobial resistance genes erm(B), lnu(C), tet(M), tet(L) and acc(6′)-aph(2”) in the genome of this strain (Table 2), which led to strain WUSP067 resistance to erythromycin, tulathromycin, lincomycin, clindamycin, doxycycline and gentamycin (Table 2). The strain resistant to three or more classes of antimicrobial agents is considered to be multidrug resistant (Schwarz et al., 2010). Thus, strain WUSP067 is multidrug resistant. To explore vehicles harbouring antimicrobial resistance genes for S. pasteurianus, the ICE and prophage were predicted. Three incomplete prophages with scores ≤ 20 were predicted without antimicrobial resistance genes. Six ICEs were predicted, and two of them contained antimicrobial resistance genes. The ICEWUSP067-1 (from E8M05_RS07995 to E8M05_RS08435) harbouring a 15-bp att sequence 5′-tcactaatatcttga-3′ in the flanking region contained genes erm(B) and acc(6′)-aph(2″) (Figure 2a). The ICEWUSP067-2 (from E8M05_RS10655 to E8M05_RS11075) harbouring a 15-bp att sequence 5′-aattgcttcccaagt-3′ in the flanking region contained genes tet(M) and tet(L) (Figure 2b).
| Classes | Antibiotics | Breakpoints for resistancea (mg/L) | MICs (mg/L) | Resistance mechanisms | Inside of MGEs |
|---|---|---|---|---|---|
| Macrolides | Erythromycin | ≥1 | >256 | erm(B) | ICEWUSP067-1 |
| Tulathromycin | ≥64 | 128 | erm(B) | ICEWUSP067-1 | |
| Lincosamides | Lincomycin | ≥1 | >256 | lnu(C) | No.2 GI |
| Clindamycin | ≥1 | >256 | lnu(C) | No.2 GI | |
| Tetracyclines | Doxycycline | ≥1 | 32 | tet(M), tet(L) | ICEWUSP067-2 |
| Aminoglycosides | Gentamycin | ≥16 | >256 | acc(6′)-aph(2″) | ICEWUSP067-1 |
- MGEs, mobile genetic elements; GI, genomic island.
- a The breakpoint for resistance to erythromycin and clindamycin was based on the CLSI for Streptococcus viridans group (Li et al., 2019). Since the breakpoint for resistance to the rest of antimicrobial agents is not available, the breakpoint for resistance to the rest of antimicrobial agents was adopted according to the previous study (Wang et al., 2021).
3.3 Strain WUSP067 contains a fucose utilization cluster sharing a high identity with that of S. suis
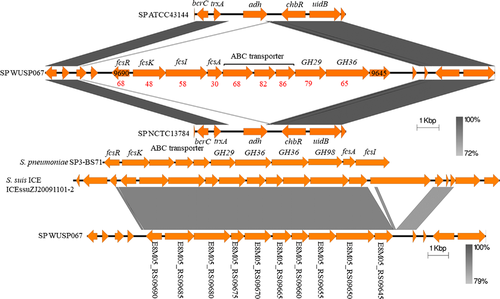
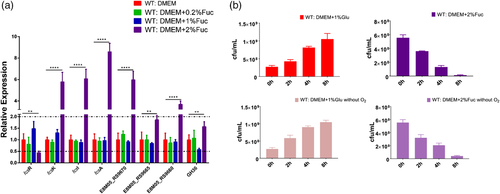
Although the genome sequence of strain WUSP067 shared high homology with that of human isolates, the comparative genome analysis showed that it contains 36 unique fragments larger than 1 kb absent in human isolates (Figure 1b and Supplementary Table S4). Notably, the 89% nucleotide sequence of No. 23 unique fragment shared 91% identity with that of an ICE ICEssuZJ20091101-2 from S. suis, another important swine bacterial pathogen. Furthermore, this unique fragment contains a putative fucose utilization cluster (from E8M05_RS09645 to E8M05_RS09690), and the amino acid sequence of proteins predicted in this cluster shared 30–79% identity with that of proteins related to fucose utilization in Streptococcus pneumoniae (Figure 3). To investigate if strain WUSP067 is responsive to fucose, strain WUSP067 was cultured in DMEM without the carbohydrate or containing fucose with a final concentration of 0.2%, 1% or 2% for 2 h. When the final concentration of fucose reached 2%, compared with the control condition, the expression of fcsR was significantly decreased, while that of the rest genes shown in Figure 4a was significantly increased. However, we found that strain WUSP067 cannot use fucose as the sole carbon source with a final concentration of 2% with or without O2 (Figure 4b).
3.4 Strain WUSP067 causes meningitis and pathological changes of brain tissue in 3-week-old ICR mice
So far, the pathogenic characteristic of S. pasteurianus is unknown. The 3-week-old ICR mice were used to investigate S. pasteurianus pathogenicity. Four groups of mice (10 mice per group) were intraperitoneally injected with strain WUSP067 with doses of 1.5×109, 3×108, 6×107 or 1.2×107 CFU per mouse. The mice began to die 2 days after the challenge, and all the mice in the highest dose challenge group died within 4 days. The meningitis symptoms, including convulsions, rigid posture or ataxia, appeared 5 days after the challenge. In total, 21 mice stayed alive on the fifth day post-infection, and 61.9% of mice (13/21) showed meningitis symptoms before death. Five dead mice were randomly selected, and S. pasteurianus can be isolated from the brain, heart, liver, spleen and lungs of these five dead mice. The LD50 value of strain WUSP067 was 4.0×107 CFU per mouse. In addition, we found that the weight of the mice infected with strain WUSP067 with doses of 6.4×107 or 1.7×106 CFU per mouse was decreased (Supplementary Figure S1). The mice in the control group injected with PBS grew normally.
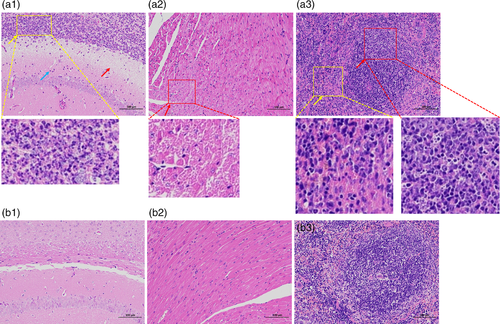
The histopathological examination was performed to clarify the pathogenicity of this bacterium further. Six mice were intraperitoneally injected with strain WUSP067 with doses of 1×107 or 1×108 CFU per mouse (three mice per dose). The brain of mice infected with strain WUSP067 showed neutrophils infiltration in the hippocampus (blue arrow), loose structure and necrosis of the corpus callosum (red arrow) and diffuse purulent lesions (yellow arrow) (Figure 5a1). All six mice showed pathological changes in their brains. The slight necrosis of cardiomyocytes was observed in the heart of mice in infection groups (red arrow) (Figure 5a2). Three of six mice showed pathological changes in hearts. The pyknosis of lymphocyte nuclei at white pulp (red arrow) and the haemorrhage at red pulp (yellow arrow) were observed in the spleen of mice in infection groups (Figure 5a3). Three of six mice showed pathological changes in spleens. The above tissues of mice in the control group injected with PBS did not find any pathological changes (Figure 5b1-3). The description of pathological changes for each mouse in infection groups is summarized in Supplementary Table S5.
3.5 The capsule is an essential virulence factor for S. pasteurianus
So far, there is no report about S. pasteurianus virulence factors. Since the capsule is an important virulence factor for many pathogenic Streptococcus species, we investigated if the capsule would also contribute to S. pasteurianus virulence. The gene-deletion mutagenesis method is a major step for studying S. pasteurianus pathogenesis. The temperature-sensitive suicide plasmids pSET4s and pSET5s have been used to construct deletion mutants in several pathogenic Streptococcus species successfully, including S. suis (Takamatsu et al., 2001b), Streptococcus equi subsp. zooepidemicus (Ma et al., 2019) and Streptococcus agalactiae (Liu et al., 2020). However, these plasmids cannot be introduced into strain WUSP067 by electrotransformation. The natural transformation is a gene horizontal transfer mechanism, which provides a valuable resource for developing genetic editing methods in Streptococcus genus (Fontaine et al., 2015; Zhu et al., 2019). The genes encoding for ComRS system involved in competence regulation were predicted in the genome of strain WUSP067: E8M05_RS00415 was predicted encoding ComR, and 15-aa peptide (ComS, MLNIFSIVLTGWWGV) was predicted on the 5′ of comR. The N-terminal cleavage is a necessary process for ComS to generate the active form of the peptide in Streptococcus genus (Fontaine et al., 2015; Zhu et al., 2019). Based on our preliminary experiments, the transformation efficiency was maximal with ComS7-15 (IVLTGWWGV), suggesting that ComS7-15 closely resembles the active form of this pheromone. A genetic editing method for S. pasteurianus was developed using ComRS natural transformation system. The linear DNA can be uptaken by naturally transformable bacteria as foreign genetic material during transformation (Zhu et al., 2019). Thus, the fusion fragment, upstream of the target gene (AB)-spc-downstream of target gene (CD), was constructed by overlapping PCR, which contains the positive selection marker spc. After transformation and spectinomycin-resistant selection, the target gene was replaced with the spc, and mutants presented a spectinomycin-resistant phenotype (Figure 6a).
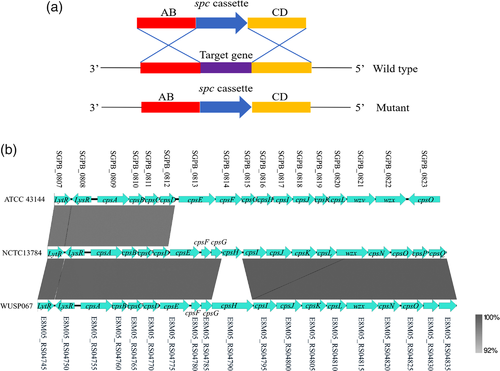
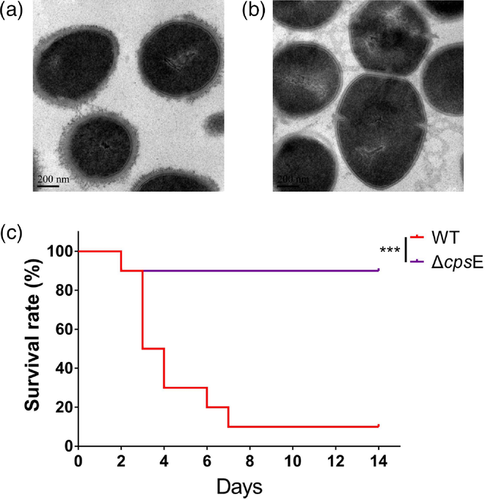
Since the deletion of gene encoding initial glycosyltransferase involved in capsule synthesis caused capsule loss in S. suis (Lakkitjaroen et al., 2014), the gene encoding initial glycosyltransferase (cpsE, Figure 6b) was selected to test whether the gene-deletion mutagenesis method works in strain WUSP067. The locus of capsule synthesis in S. pasteurianus strains WUSP067, NCTC 13784 and ATCC 43144 is located downstream of gene deoD encoding the purine-nucleoside phosphorylase. E8M05_RS04775 is the first glycosyltransferase in the cps locus in WUSP067. The deletion mutant of cpsE (ΔcpsE) was successfully constructed. The TEM analysis showed that wild-type strain was encapsulated, while the capsule of ΔcpsE was lost (Figure 7a and b). Furthermore, the virulence of ΔcpsE was evaluated in the above mouse infection model. There is a significant difference in survival curves between mice infected with the wild-type strain and ΔcpsE (Figure 7c). Mice infected with wild-type strain with the dose of 3×108 CFU per mouse had a 10% survival rate on the 14th day post-infection, while mice infected with ΔcpsE with the same dose had a 90% survival rate at the same time point. This finding confirmed that the capsule was an essential virulence factor of S. pasteurianus.
4 DISCUSSION
S. pasteurianus, an underreported opportunistic pathogen, is considered an increasingly recognized cause of meningitis and bacteremia in many animals and humans worldwide (Jans et al., 2015; Klatte et al., 2012; Li et al., 2019). We first isolated an S. pasteurianus strain from a piglet's brain with meningitis. The strain WUSP067 caused meningitis and pathological changes of brain tissue in the mouse infection model. Thus, S. pasteurianus can also be considered as an opportunistic pathogen for pigs.
Strain WUSP067 is multidrug-resistant. The multidrug-resistant bacteria could possess a competitive advantage over other microbiota to preferentially colonize the host when antimicrobial selective pressures exist. Li et al. reported that 88.9% (40/45) S. pasteurianus strains isolated from patients were both erythromycin and clindamycin resistant, and 97.5% (39/40) of them carried erm(B) gene. In addition, they showed that 95.6% of strains (43/45) were tetracycline-resistant, and most strains carried tet(L), tet(O) or tet(M), alone or in combination (Li et al., 2019). However, how the spread of antibiotic resistance genes occurs is unknown. In this study, we showed that strain harboured five antimicrobial resistance genes erm(B), lnu(C), tet(M), tet(L) and acc(6′)-aph(2”), leading to this strain resistance to erythromycin, tulathromycin, lincomycin, clindamycin, doxycycline and gentamycin. Four of them, erm(B), tet(M), tet(L) and acc(6′)-aph(2″), are present in two ICEs (ICEWUSP067-1 and ICEWUSP067-2). Thus, ICEs are vehicles harbouring antimicrobial resistance genes, contributing to the spread of antibiotic resistance genes in S. pasteurianus.
Although S. pasteurianus is considered as an increasingly recognized pathogen for animals and humans, the aetiology of infection needs further study. Identifying the reservoir niches of pathogens is essential for understanding the pathogenic characteristics of pathogens and the prevention and control of diseases caused by pathogens. In this study, we found that the nucleic acid of S. pasteurianus can be detected in tonsils and hilar lymph nodes from healthy pigs, suggesting that these tissues could be the reservoir niches for this bacterium. The detection rate of S. pasteurianus in hilar lymph nodes (52%) was higher than that in tonsils (12%). Further studies are needed to determine if this is related to the regions where samples were collected or tissues themselves. The pig tonsil is one of reservoir niches for S. suis (Swildens et al., 2007; Vötsch et al., 2018), and S. suis from healthy pig tonsils is considered as one of the important infection sources for pigs and humans. Whether S. pasteurianus in healthy pig tonsils and hilar lymph nodes is an infection source needs further study. Hatrongjit et al. reported that 8.6% (38/442) and 10.2% (45/442) of streptococcal isolates from seven tertiary hospitals in northeast Thailand were identified as S. suis and S. pasteurianus, respectively. The authors used gene SGPB0680 (E8M05_RS04035 in strain WUSP067) to identify S. pasteurianus, which was consistent with the results of API 20 Strep and conventional biochemical tests (Hatrongjit et al., 2017). In the present study, in addition to gene SGPB0680, two S. pasteurianus-specific genes (E8M05_RS05155 and E8M05_RS06300 in strain WUSP067) were also included to identify S. pasteurianus from pig tonsils and hilar lymph nodes. We found that there were two tonsils with the presence of only SGPB0680. The sodA was amplified from these two tonsils, and their sodAint sequence (from positions 25 to 510) shared 96.09% and 96.91% with that of type train NCTC 13784, respectively, below the standard of ≥ 98.9% to denote them as S. pasteurianus (Poyart et al., 2002). Thus, samples with the presence of all three genes were judged to be positive for S. pasteurianus.
Fucose, an L-configuration sugar, is present abundantly in the mammalian gastrointestinal tract (Pickard & Chervonsky, 2015). The enterohaemorrhagic Escherichia coli (EHEC) sensed fucose, a host-derived signal made available by Bacteroides thetaiotaomicron, by a two-component signal transduction system FusKR to regulate EHEC pathogenicity and metabolism (Pacheco et al., 2012). Campylobacter jejuni containing a genomic island required for fucose utilization allowed for using fucose as a substrate for growth; mutants with the deficiency of fucose uptake demonstrated a competitive disadvantage in vivo (Stahl et al., 2011). S. pneumoniae strain TIGR4 cannot use fucose as sole carbon, but the gene expression of fucose utilization operon was induced by fucose (Chan et al., 2003). Deletion of the whole operon attenuated S. pneumoniae (Embry et al., 2007). Strain WUSP067 contains a fucose utilization cluster sharing a high identity with that of ICEssuZJ20091101-2 from S. suis. The locus of strain WUSP067 genome near the fucose utilization cluster only contains two genes encoding putative transposases. It does not have genes encoding for conjugation machinery, while ICEssuZJ20091101-2 belongs to tandem mobile genetic elements (ICE-ICE) containing genes encoding integrases and conjugation machinery. Since S. suis can be isolated from pig tonsils and lymph nodes, these two pathogens may have the chance to contact and exchange genes. Indeed, three hilar lymph nodes were positive for these two bacteria. In addition, this strain may have acquired the fucose utilization cluster via its natural transformation. Like S. pneumoniae strain TIGR4, WUSP067 is able to respond to fucose, but it cannot use fucose as the sole carbon source. It merits further study concerning the function of the fucose utilization cluster in S. pasteurianus pathogenicity and metabolism.
The infection model is crucial for clarifying the pathogenic characteristics of pathogens. Strain WUSP067 was isolated from the brain of a newly weaned piglet with meningitis, so 3-week-old newly weaned mice were used. In this infection model, 61.9% of mice (13/21) showed meningitis symptoms before death, and all mice for the histopathological examination showed pathological changes in brains. In our preliminary infection experiments, the adult mice infected with WUSP067 also showed meningitis symptoms, but they were less susceptible than 3-week-old newly weaned mice. Thus, the stresses, such as weaning, may increase the susceptibility to S. pasteurianus. Ducklings infected with S. pasteurianus showed typical neurological symptoms, severe congestion of the meninges, and necroptosis of splenic macrophages (Li et al., 2013; Li, Liu, et al., 2017). In calves, the major pathological changes of the central nervous system were marked suppurative-meningitis with inflammatory cell infiltration, forming diffuse micro-abscesses (Trotta et al., 2019).
In addition to plasmids pSET4s and pSET5s for generating deletion mutants in many Streptococcus species, plasmids pSET1 (Takamatsu et al., 2001a), pSET2 (Takamatsu et al., 2001a), pSET3 (Takamatsu et al., 2001a) and pAT18 (Li et al., 2010) for constructing complemented strains cannot be introduced into WUSP067 either by electrotransformation or natural transformation. So far, only the linear DNA fragment can be uptaken by natural transformation in this strain. Therefore, strain WUSP067 may harbour a particular defence mechanism against foreign DNA invasion. CRISPR-Cas and RM systems are defence tools against foreign DNA in bacteria (Oliveira et al., 2016; Terns & Terns, 2011). There were two predicted CRISPR/Cas systems (Supplementary Table S6), and no RM system was predicted in WUSP067. If the CRISPR-Cas systems link with S. pasteurianus low transformation frequency needs further study. Our previous study found that some virulent strains had lower transformation frequencies, and some avirulent strains had higher transformation frequencies in S. suis (Zhu et al., 2019). Further study is needed to find out if the transformation frequency links with S. pasteurianus virulence. By natural transformation, the capsule deficient mutant was successfully generated. The virulence of the mutant was significantly attenuated in the mouse infection model, confirming that capsule is an essential virulence factor for S. pasteurianus. The capsule cluster contains 19, 17 and 19 genes in strains WUSP067, ATCC 43144 and NCTC 13784, respectively. Only gene cpsH is different in capsule cluster between strains WUSP067 and NCTC 13784, while genes from cpsE to cpsO in strain ATCC 43144 shared no identity with those in strains WUSP067 and NCTC 13784 (Figure 6b). It would be interesting to investigate if there are different serotypes based on the capsule and if the virulence of different serotypes is different in the future.
In summary, we demonstrated that S. pasteurianus is an opportunistic pathogen for pigs, and the tonsils and hilar lymph nodes of healthy pigs may be reservoir niches for this bacterium. Furthermore, we showed that ICEs are vehicles for harbouring antimicrobial resistance genes conferring resistance to host strain. Using the 3-week-old newly weaned ICR mice as the infection model, we revealed that the infected mice showed severe signs of meningitis and pathological changes in brains, and the capsule was an essential virulence factor for S. pasteurianus. In-depth studies of the pathogenic mechanism of this underreported pathogen using tools established in this study are needed.
ACKNOWLEDGEMENTS
This work was supported by the Open Project Program of Jiangsu Key Laboratory of Zoonosis under Grant No. R2103, and the Open Project Program of Engineering Research Center for the Prevention and Control of Animal Original Zoonosis, Fujian Province University under Grant No. 2021ZW001.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICAL APPROVAL
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to, and the appropriate ethical review committee approval has been received. The experiments were performed in the Laboratory Animal Center of Nanjing Agricultural University approved by the Laboratory Animal Monitoring Committee of Jiangsu Province (Permit number: SYXK (Su) 2017-0007). The experiments were performed strictly according to our institutional guidelines, and all efforts were made to minimize suffering.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in the supplementary material of this article.




