Is integrated taxonomy useful to study diversity and ecology? An example from crustacean zooplankton at the Long-Term ecological research site MareChiara (LTER-MC)
Abstract
Molecular tools increasingly refine and improve the identification of zooplankton organisms based on phenotypic features, providing a more robust and comprehensive species description. Integration of data helps revealing the hidden diversity of zooplankton and facilitating the detection of rare and non-indigenous species. This approach, merging morphological characters and a diagnostic marker for specific identification, such as the mitochondrial cytochrome c oxidase I (COI), is here used to characterize key taxa from the zooplankton assemblage of the Gulf of Naples at the Long-Term Ecological Research site MareChiara (LTER-MC) (Central Tyrrhenian Sea, Western Mediterranean Sea). Zooplankton biodiversity assessment using integrated taxonomy was focused on selected crustacean groups: cyclopod copepods (Agetus typicus, Oithona plumifera, Oncaea mediterranea, Oncaea scottodicarloi); newly records of cladocerans (Evadne nordmanni), euphausiids (Euphausia krohnii, Nematoscelis megalops, Nyctiphanes spp.) and sergestids (Lucifer typus), with the aim to boost the knowledge of real zooplankton biodiversity. The results of our investigation provide new high-quality molecular references of the analysed taxa and contribute to unveiling the genetic diversification of zooplankton species and their relevant ecological significance for Mediterranean coastal waters.
1 INTRODUCTION
Zooplankton time-series are fundamental in the assessment of the state and the health of marine ecosystems (Boero et al., 2015; O'Brien et al., 2017; Ratnarajah et al., 2023). Zooplankton are indeed recognised among the essential biological variable in the framework of the 2022 GCOS Implementation Plan (https://gcos.wmo.int/en/publications/gcos-implementation-plan2022) and as an indicator of the good environmental status in the EU Marine Strategy Framework Directive (2008/56/EC https://ec.europa.eu/environment/marine/good-environmental-status/index_en.htm).
Zooplankton assemblages include representatives of the entire spectrum of taxa, spanning from protozoans to chordates (Bucklin et al., 2021; Bucklin, Nishida, et al., 2010). These organisms are central components of a holistic ecosystem assessment due to their intermediary role in the food web, linking lower with higher trophic levels. As a result, they incorporate the inherent properties and variations occurring at all levels of marine ecosystems, their changes in communities being important indicators of both the overall ecosystem health and global impacts.
Notwithstanding its importance, zooplankton diversity is still particularly challenging to assess. At present, this task is performed mostly based on the morphological identification of species. In monitoring programs, zooplankton taxa are generally identified only to the class or family level (O'Brien et al., 2017), and only well-known and dominant species (mainly copepods) are identified accurately, whereas other taxa, including rare ones, are often underestimated (Laakmann et al., 2020). The taxonomic identification of zooplanktonic organisms based on morphological characters requires a high level of specialization (Di Capua et al., 2022; Zingone et al., 2019), but the number of expert taxonomists is rapidly declining (Laakmann et al., 2020). Moreover, the occurrence of numerous cryptic species and species complexes, together with limited diagnostic characters for meroplanktonic species, further hinders the assessment of zooplankton biodiversity and biogeography, with frequent species misidentifications (Di Capua et al., 2022; Laakmann et al., 2020). In the last two decades, the use of molecular taxonomy in zooplankton research has led to a revolution in species identification and characterization, as well as in the assessment of biodiversity (Djurhuus et al., 2018). The mitochondrial cytochrome C oxidase I (COI) gene has proved successful in the identification of zooplanktonic organisms and is considered a reference gene for taxonomic purposes (Blanco-Bercial et al., 2014; Bucklin, Hopcroft, et al., 2010; Bucklin, Nishida, et al., 2010; Laakmann et al., 2020). The creation of robust reference libraries, however, requires prior accurate morphological identification to align phenotypic and genotypic information (Bucklin et al., 2021).
Globally, Copepoda dominate marine zooplankton (Boxshall & Halsey, 2004), being highly diversified in morphology, physiology, behavior, habitat preference, geographic distribution and climatic tolerance (Uttieri, 2018). They represent up to 90%–97% of the marine zooplankton biomass (Bradford-Grieve et al., 1999), living in any type of environment from deep-sea trenches to neuston (Huys & Boxshall, 1991), and from polar waters to hydrothermal vents (Walter & Boxshall, 2022).
Cladocera are small and cosmopolitan crustaceans seasonally abundant in coastal waters (Rivier, 1998). Out of the approximately 620 known species (Forró et al., 2008), only eight are truly marine, an unbalance that can be considered as “curious” (Durbin et al., 2008). Nevertheless, their broad distribution makes them eligible for phylogeographic and population genetic studies (Durbin et al., 2008). Marine cladocerans are restricted to coastal waters, where they make up a significant part of the zooplanktonic community at given periods, and in Gulf of Naples they dominate in summer (Mazzocchi et al., 2023).
Among Malacostraca, Euphasiacea and Sergestoidea are of particular importance in the pelagic realm. In the Mediterranean Sea, only thirteen species referred to seven genera of Euphausiacea are reported (Costanzo & Guglielmo, 1976; Guglielmo, 2010), while systematics and ecology are well studied mainly in the North Atlantic and South Pacific Oceans (Bucklin et al., 2007; Gibbons, 1997; Gibbons et al., 1995; Mauchline, 1980). Decapods of the family Luciferidae (superfamily Sergestoidea) are a typical component of tropical and subtropical epipelagic systems (Omori, 1992). They have rarely been reported from the Mediterranean Sea, making their study in this basin particularly challenging (Galil & Shlagman, 2010).
More than 200 zooplanktonic taxa have been regularly identified through diagnostic morphological characters at the Long-Term Ecological Research station MareChiara (LTER-MC) in the Gulf of Naples (GoN; Central Tyrrhenian Sea, Western Mediterranean Sea) from 1984 to 2015 (Mazzocchi et al., 2023). In a previous paper, phenotype-based approaches were combined with molecular ones to identify key calanoid copepod species collected at LTER-MC, revealing the occurrence of cryptic species and the biogeographic connections among different populations (Di Capua et al., 2022). The same methodological framework is here extended to other relevant components of the LTER-MC zooplankton assemblage, focusing on key target crustacean taxa (Copepoda, Cladocera, Malacostraca) with a pivotal role in the pelagic ecosystems, not only at the investigated station but also in neritic areas worldwide (Castellani & Edwards, 2017). Starting from the multidecadal knowledge of the zooplankton assemblage at LTER-MC, in this study we complement the routine morphological taxonomic identification of zooplanktonic organisms with DNA barcoding to disentangle the identification of key target species: (i) regularly found copepods that require highly laborious taxonomic identification (Agetus typicus, Oithona plumifera, Oncaea mediterranea, Oncaea scottodicarloi); (ii) a recently recorded cladoceran (Evadne nordmanni), never observed before at LTER-MC; and (iii) malacostraca often forming dense and abundant swarms at LTER-MC, but not easily identified, in particular during their life stages (the euphausiids Euphausia krohnii, Nematoscelis megalops, Nyctiphanes spp., and the sergestid Lucifer typus). The study is aimed at expanding the current knowledge of the biodiversity of zooplanktonic taxa using integrated taxonomy. This lays the foundation and contributes to DNA barcoding libraries for Mediterranean coastal areas, starting from the GoN due to its high representativeness, by providing new high quality and validated reference sequences. From a broader perspective, by complementing taxonomic description with biological and ecological traits of the investigated taxa we aim at drawing a more comprehensive picture of their role in different marine environments, as well as of their phylogeographic connections in Mediterranean coastal waters.
2 MATERIALS AND METHODS
2.1 Sampling
Zooplankton samples were collected in the GoN at the coastal LTER-MC site. A WP2 plankton net (mouth diameter: 57 cm; mesh aperture width: 200 μm) was towed vertically from −50 m depth to the surface at low speed (0.7–1.0 m s−1). All sampling activities were performed in the framework of the LTER-MC activities and of the Naples Ecological REsearch and Augmented ocean observation (NEREA) project (https://www.nerea-observatory.org/), an augmented observatory including “omic approaches”. Upon collection, the live material was brought within 2 h to the laboratory, where it was filtered on a 200 μm nitex filter, preserved in 95% ethanol and stored in the dark at 4°C. After 24 h, the ethanol was replaced to remove seawater excess.
2.2 Morphological analyses
Individual organisms were sorted out from LTER-MC zooplankton samples under a Leica M165C stereomicroscope. For copepods and cladocerans, the identification was carried out at species level on adult females using diagnostic morphological characters and classification according to Boltovskoy (1999), Razouls et al. (2005-2022), and Trégouboff and Rose (1957), checking original descriptions and revisions, when relevant.
Malacostraca larvae, euphausiid eggs and larval stages were identified using the taxonomic keys reported by Castellani and Edwards (2017), while for Sergestoidea the work by Dakin and Colefax (1940) was used as reference.
The taxonomic classification and nomenclature of all taxon names are in agreement with the World Register of Marine Species (https://www.marinespecies.org/wormsliterature.php).
2.3 Selected key taxa
2.3.1 Copepoda, Cyclopoida
Eight Cyclopoida specimens referred to four species from two orders were selected. Oncaea species are very abundant and regularly collected throughout the years at the LTER-MC site (Di Capua et al., 2017). Oncaea scottodicarloi was described by Heron and Bradford-Grieve (1995) as a distinct species based on specimens collected in the GoN, while Oncaea mediterranea was discovered for the first time in the Mediterranean Sea by Claus (1863). Oithona plumifera is one of the most abundant of the eight Oithona species at the LTER-MC site, while Agetus typicus is a rare species regularly observed since 1984 (Di Capua & Mazzocchi, 2021).
2.3.2 Cladocera
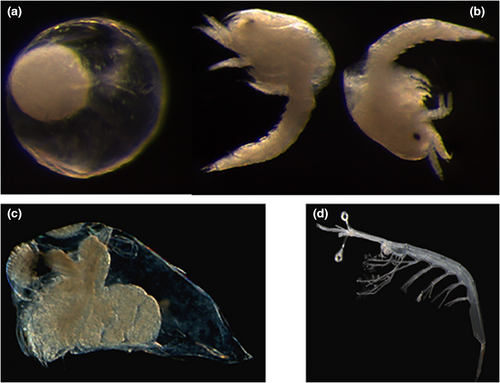
At LTER-MC site, five species (Evadne spinifera, Pseudevadne tergestina, Penilia avirostris, Podon intermedius and Pleopis polyphaemoides) are regularly recorded with a clear seasonal succession among them from April to August (Montalbano, 2021). In addition, the present study includes a species morphologically identified as Evadne nordmanni, which was recorded for the first time at LTER-MC in April 2021 (Figure 1c).
2.3.3 Malacostraca
Euphausiacea and Sergestoidea swarms are recorded at the LTER-MC site occasionally, but they are identified only at high taxonomic level. Usually, eggs and different larval stages are collected in March–April. In January 2021, a swarm of eggs and calyptopis of Euphausiidae were collected, with more than 41 ind. m−3 (Figure 1a,b). Two eggs and nine calyptopis stages were selected for this study, and identified morphologically.
Among decapod larvae, a zoea of the suborder Dendrobranchiata, superfamily Sergestoidea (Figure 1d) was morphologically identified as a zoea of Luciferidae (Dakin & Colefax, 1940).
2.4 Molecular analyses
DNA extraction, purification, and PCR were performed as reported by Di Capua et al. (2022). Folmer et al. (1994) primers pairs (LCO1490:5′-GGTCAACAAATCATAAAGATATTGG-3 HCO2198: 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) were used in PCR to amplify the COI gene of crustaceans, considered a universal marker for marine zooplankton (Bucklin et al., 2011, 2021; Bucklin, Hopcroft, et al., 2010).
Consensus sequences were generated with BioEdit Sequence Alignment Editor (Hall, 1999) and unique sequences (collapse of identical sequences) were obtained with mothur v.1.44.3 (Schloss et al., 2009). Identification by similarity was performed blasting our sequences against the Barcode of Life Data Systems (BOLD) (Meiklejohn et al., 2019; Ratnasingham & Hebert, 2007) and GenBank databases (http://www.ncbi.nlm.nih.gov). Moreover, identification by generation of and placement in a tree (phylogenetic approach) was performed by downloading reference sequences from GenBank and/or BOLD (Meiklejohn et al., 2019; Ratnasingham & Hebert, 2007). Such reference sequences were filtered to remove low quality sequences (barcode <400 base pairs; bp), not identified to species level, or presenting ambiguities (as in Di Capua et al., 2022) and combined with our Sanger sequences. The software MAFFT (Katoh et al., 2018; Kuraku et al., 2013) was used to perform an alignment of the total sequence data, which was subsequently checked in Sea View v4.0 (Gouy et al., 2010). Maximum Likelihood (ML) trees (GTR model) were constructed with Fastree (Price et al., 2010) and visualized in the iTOL (Interactive Tree Of Life) software (Letunic & Bork, 2019).
In the case of Cyclopoida, 359 COI reference sequences, referred to the three analyzed copepods (Oncaea, Oithona and Agetus), were downloaded from BOLD and, subsequently, filtered to remove low quality data. Nine insect sequences (four species) where chosen as outgroups (Anopheles pristinus accession numbers GU989357, GU989358, GU989348; Gressittacantha terranova accession numbers HM461319, HM461301, HM461312, HM461287; Lepicerus inaequalis accession number KJ871320; and Mycetaulus bipunctatus accession number KR436825).
For Cladocera, 224 COI reference sequences were downloaded from BOLD and were filtered as above. The alignment included 92 filtered data from BOLD, the E. spinifera sequences from the GoN and one sequence of a calanoid copepod species (Calanus helgolandicus accession number JX995315.1) as an outgroup (Di Capua et al., 2022).
In Euphausiacea trees, 1,362 COI reference sequences were downloaded from BOLD and GenBank, and were filtered as above, generating a final dataset including 95 COI data. The multialignment included filtered data from BOLD and GenBank, our euphausiacea COI sequences and the outgroup, represented by four Stomatopoda sequences (BOLD: Oratosquilla oratoria accession number NC014342; GenBank: Harpiosquilla harpax accession number NC006916, Squilla empusa accession number NC007444 and Squilla mantis accession number NC006081).
Phylogenetic networks were generated for E. nordmanni and Nyctiphanes genus. Haplotype lists were generated with DnaSP (Rozas et al., 2017), then Median-Joining (MJ) haplotype networks were inferred with Network-fluxus 10 using default parameters.
3 RESULTS
3.1 Morphological and molecular identification
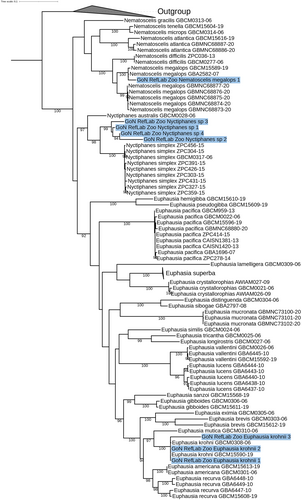
The COI sequences generated were longer than 400 bp. Analysis by similarity confirmed morphological identification at species level for copepods and cladocerans with high similarity (98–100%) (Table 1). The sequences generated from eleven specimens morphologically identified as eggs and calyptopis of Euphausiacea were assigned to the two species Nematoscelis megalops (99% of similarity) and Euphausia krohnii (100% of similarity). The rest of the sequences were assigned to the genus Nyctiphanes (92% of similarity), corresponding to a reference of N. simplex from the Pacific Ocean, since no molecular references for this taxon are available from the Mediterranean Sea. The larva of Sergestoidea was morphologically and molecularly identified as Luciferidae, according to the low similarity (94%) with a reference of Lucifer typus, a species reported also from the Mediterranean Sea (Galil & Shlagman, 2010), but lacking of molecular reference from this area.
| Taxon | Genus | Species | Authors | WoRMS (AphiaID) | Morphoplogical identification | N° specimens analized | Code | N° of sequences | Molecular identification | % NCBI or BOLD |
|---|---|---|---|---|---|---|---|---|---|---|
| Cyclopoida | Oncaea | O. scottodicarloi | Heron & Bradford-Grieve, 1995 | 128,950 | O. scottidicarloi | 5 | GoN_RefLab_Zoo_Oncaea scottodicarloi_1–3 | 3 | O. scottidicarloi | 100 |
| “ | O. mediterranea | (Claus, 1863) | 128,939 | O. mediterranea | 1 | GoN_RefLab_Zoo_Oncaea mediterranea_1 | 1 | O. mediterranea | 98 | |
| Oithona | O. plumifera | Baird, 1843 | 106,652 | O. plumifera | 1 | GoN_RefLab_Zoo_Oithona plumifera_1 | 1 | O. plumifera | 99 | |
| Agetus | A. typicus | Krøyer, 1849 | 852,051 | A. typicus | 1 | GoN_RefLab_Zoo_Agetus typicus_1 | 1 | at order level | 93 | |
| Cladocera | Evadne | E. nordmanni | Lovén, 1836 | 106,273 | E. nordmanni | 4 | GoN_RefLab_Zoo_Evadne nordmanni_1–4 | 4 | E. nordmanni | 100 |
| Euphausiacea | Euphausia | E. krohnii | (Brandt, 1851) | 110,687 | Euphasiacea 2 eggs and 2 calyptosis | 4 | GoN_RefLab_Zoo_Euphausia krohnii_1–3 | 3 | E. krohnii | 100 |
| Nematoscelis | N. megalops | G.O. Sars, 1883 | 110,695 | Euphasiacea 2 calyptosis | 2 | GoN_RefLab_Zoo_Nematoscelis megalops_1 | 1 | N. megalops | 99 | |
| Nyctiphanes | Nyctiphanes sp. | Hansen, 1911 | 237,864 | Euphasiacea 5 calyptosis | 5 | GoN_RefLab_Zoo_Nyctiphanes sp._1–4 | 4 | N. simplex | 92 | |
| Decapoda | Lucifer | L. typus | H. Milne Edwards, 1837 | 883,865 | Luciferidae | 1 | GoN_RefLab_Zoo_Lucifer_sp._1 | 1 | L. typus | 94 |
3.2 Copepoda, Cyclopoida
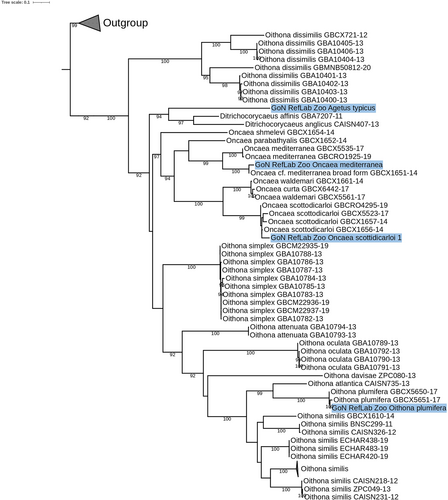
In the COI ML tree, all epipelagic species of Oncaea sensu stricto clustered together (Figure 2) in a highly supported clade (96%). Our sequence of O. mediterranea showed 100% identity with the sequence of Oncaea cf. mediterranea (broad form, GBCX1651-14), and indeed robustly clustered with two other references of O. mediterranea from the Eastern Mediterranean Sea. In the O. scottodicarloi clade our sequences robustly clustered with samples from Greece (99% bootstrap). Seven epipelagic species of Oithona generated several clades, while the sequences from our specimens identified morphologically as O. plumifera clustered (100% bootstrap) with other two O. plumifera references from Villefranche sur Mer (Figure 2). Our new reference of Agetus typicus grouped with the only two references available for corycaeids of the genus Ditrichocorycaeus (Figure 2).
3.3 Cladocera
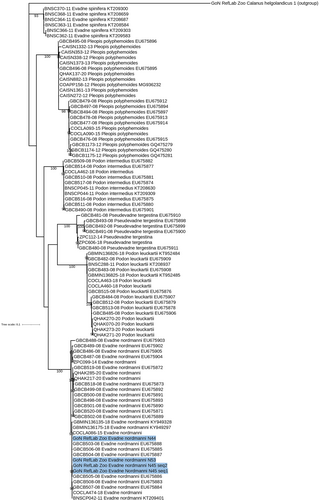
The morphological identification of four parthenogenetic females of Evadne nordmanni was confirmed by molecular analyses (100% of similarity with EU675885.1), and the placement in the tree showed our new sequences from the Mediterranean Sea clustering into a sister clade with other references from the Baltic Sea (Figure 3). The two globally distributed Evadne species, namely E. spinifera and E. nordmanni, grouped in two well supported and separated clades (99.9% bootstrap). Finally, the haplotype network of E. nordmanni including 33 COI sequences generated 22 haplotypes with high haplotype diversity (h = 0.97) (Figure 4). The specimens from the GoN produced two haplotypes (H7 and H14) related to several haplotypes with a clear geographic signal (Figure 4).
3.4 Euphausiacea
New reference sequences from the Mediterranean Sea were obtained for three genera: Euphausia, Nematoscelis and Nyctiphanes. The COI ML tree showed our first four COI sequence for Nyctiphanes sp. calyptopis clustering all together within the Nyctiphanes clade as sister species of N. australis (Figure 5). Our calyptopis sequence was identified as Nematoscelis megalops (bootstrap 100%), clustering in the clade including other reference of this species (Figure 5). The genus Euphasia showed a big, highly supported clade (93%) including twenty species. The sequences generated from two eggs and one calyptopis were placed in the clade (100% bootstrap) with other two references of E. krohnii from the Atlantic and Pacific Oceans.
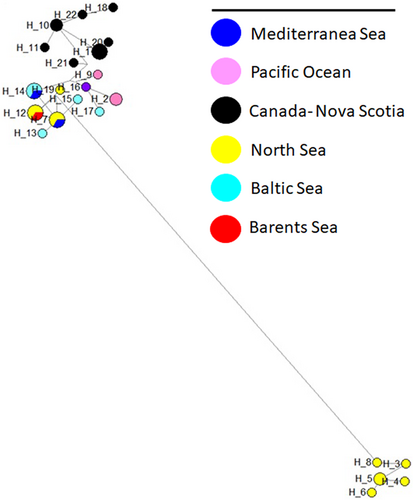
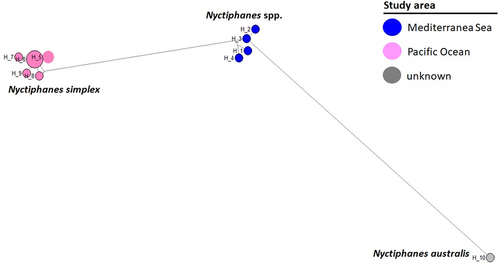
The network analysis of the genus Nyctiphanes, using the fourteen available sequences, showed ten haplotypes and high haplotype diversity (h = 0.92). The four specimens from the GoN (Mediterranean Sea) generated different haplotypes (H1-H4) and showed a clear separation from other species (Figure 6).
4 DISCUSSION
The integrated morphological and molecular analysis of a set of crustacean species of the LTER-MC zooplankton community, where they have been observed over the years, allowed to link the individual identification (α taxonomy) to a wider Ω taxonomy framework. Such integration is pivotal not only for the knowledge of the species itself (Tanduo et al., 2021, 2022) but also to assess the diversity of a system and the phylogeographic relationships among distant populations (Di Capua et al., 2022; Goetze & Ohman, 2010; Pereira et al., 2017). The crucial aspect is represented by the availability of robust, verified DNA barcode references, which guarantee the perfect correspondence between the phenotypic and molecular identifications.
Indeed, molecular data associated with long-term quali-quantitative datasets linking taxonomic, ecological and molecular approaches to the study of zooplankton taxa, are also crucial for the development of robust metagenomic and metabarcoding assessments.
At LTER sites, such as LTER-MC, expertise and knowledge of morphology-based zooplankton taxonomy built and maintained across years are now leading to the generation of high-quality reference sequences associated with morphological features, to enhance molecular taxonomy and molecular ecology studies, paving the way for an augmented omic-observatory.
Planktonic Cyclopoida (Oithonidae, Oncaeidae and Corycaeidae) are one of the most important and abundant copepod groups in oligotrophic waters (Gallienne & Robins, 2001; Medellín-Mora et al., 2021). Cyclopoida can be distinguished at species level based on detailed morphological identification (Böttger-Schnack & Machida, 2011; Heron & Frost, 2000; Paffenhofer, 1993), but validated references are almost absent in public molecular datasets. Our sequences obtained from the GoN specimes showed that COI barcoding can accurately identify also cyclopoids at the species level (98%–100% similarity), as observed for calanoid species (Di Capua et al., 2022). COI barcoding can accurately identify Oncaea species, as discussed in Di Capua et al. (2017); in addition, a new reference of O. mediterranea was obtained from a site close to its type locality (Messina; Tyrrhenian Sea). Based on COI barcoding, Oithona plumifera from the GoN is genetically related to the other populations from the Western Mediterranean Sea, also confirming its close relationships with O. atlantica and O. similis (Cornils et al., 2017). The O. plumifera sequences from the GoN group with other references from the Mediterranean Sea, suggesting a distinct molecular lineage for this basin, without indicating crypticity as shown for O. similis (Cornils et al., 2017).
The taxonomic and molecular classification of Brachiopoda is under revision (Durbin et al., 2008; Onbé, 1999; Richter et al., 2001), and our tree represents the first overview of the evolutionary relationships including Mediterranean marine cladocerans. Our COI fragments do not show monophyly at the genus level, but a good resolution at the species level, in agreement with Durbin et al. (2008). We provide the first DNA barcode of Evadne nordmanni, which also represents the first reference for the Mediterranean Sea. In the North Adriatic Sea, E. nordmanni is regularly abundant in May and then less abundant in September (Aubry et al., 2012). The species has also been reported occasionally in two coastal sites of the Tyrrhenian Sea (Margiotta et al., 2020; Umani et al., 2010). Compared to the other cladocerans typically recorded at the LTER-MC in summer (Mazzocchi et al., 2023), E. nordmanni appears in early spring at lower temperatures and salinities.
High haplotype diversity and high number of haplotypes spread from NE Pacific, SW Pacific to NE Atlantic, including the Baltic Sea, support the hypothesis of rapid colonization of the world ocean by this species (Durbin et al., 2008). However, the actual distribution of E. nordmanni may go unreported because of the close similarity of this species to the congener E. spinifera, the two being difficult to tell apart based on morphological characters only. The knowledge of the distribution of marine cladocerans is incomplete, particularly in the Mediterranean Sea. Our haplotypes from the GoN are similar to those of the Baltic Sea and the North Sea, supporting E. nordmanni as an ideal candidate for transport in ballast water and in ballast tank sediments by transoceanic ships (Cristescu & Hebert, 2002; Durbin et al., 2008) thanks to their biological and ecological traits, including euryhalinity and temperature tolerance (Rivier, 1998), as well as long survival as resting eggs (Egloff et al., 1997; MacIsaac et al., 1999; Möllmann et al., 2002).
The identification of planktonic larval stages is difficult or impossible at species and even at genus level: fine taxonomic identification is only possible by molecular tools (Brandner et al., 2017; Di Capua, Micarelli, et al., 2021, Di Capua, Piredda, et al., 2021; Walczyńska et al., 2019). In many marine species, the presence of pelagic larval stages, together with the absence of obvious distribution barriers, suggest a high level of gene flow (Palumbi, 2003). The distribution of meroplankton taxa is connected to the characteristics of the local benthic communities according to hydrodynamic and environmental conditions (Morgan, 2001). Meroplankters are important contributors to the zooplankton communities, but they are very often underestimated during routine time-series analyses (Lindeque et al., 1999). Recently, environmental DNA approaches applied at LTER-MC have revealed the high and hidden diversity of larval stages (Di Capua, Piredda, et al., 2021). Molecular data generated from crustacean larval stages (eggs and larvae) have produced four new references of euphausiids and sergestids from the Mediterranean Sea. The surface and vertical currents in the GoN may favour the entrance of offshore species, including those living in proximity to the submarine canyons engraving the bottom of the GoN (Cianelli et al., 2015). Our data suggest the presence of new species within the Nyctiphanes genus from the Mediterranean Sea, but additional morphological and molecular studies on adults are needed to disentangle this issue.
The analysis of zooplankton species using the COI gene builds an informative framework to identify and explore priority issues in the diversity, structure and functioning of zooplankton communities. Long-term series are essential baselines to evaluate the evolution of biodiversity in terms of species composition and their relative abundances. In this view, integrated taxonomy supports a wider appreciation of biodiversity, and helps to evaluate our impacts on biodiversity and the efficacy of measures aimed at reducing them.
AUTHOR CONTRIBUTIONS
IDC conception and design; IDC, RD, CM, AM acquisition of data; IDC, RP analysis and interpretation of data; IDC, MU, NB drafting the manuscript; all authors revising it critically.
ACKNOWLEDGMENTS
We thank all LTER-MC and NEREA teams and the crew of the R/V Vettoria for assistance during the work at sea. We are grateful to Elvira Mauriello and Raimondo Pannone of the Molecular Biology and Sequencing Service for technical assistance. We thank the WGIMT and WGZE of the International Council for the Exploration of the Sea (ICES) for facilitating this research. This study benefitted from collaboration with members of the Scientific Committee on Oceanic Research (SCOR) Working Group 157 (MetaZooGene). IDC and MU acknowledge the support of NBFC (National Biodiversity Future Center) to Stazione Zoologica Anton Dohrn, funded by the Italian Ministry of University and Research, PNRR, Missione 4 Componente 2, “Dalla ricerca all'impresa”, Investimento 1.4, Project CN00000033. The authors would like to thank the editor-in-chief, handling editor and anonymous reviewers for their helpful comments and suggestions on the manuscript.
FUNDING INFORMATION
The research program Long-Term Ecological Research MareChiara (LTER-MC) (LTER_EU_IT_061) was supported by Stazione Zoologica Anton Dohrn (SZN).
CONFLICT OF INTEREST STATEMENT
The authors have declared no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The sequences presented in this study are deposited online on NCBI (https://www.ncbi.nlm.nih.gov/) with accession numbers (OQ785653-OQ785662).




