Phylotranscriptomic insights into Asteraceae diversity, polyploidy, and morphological innovation
Edited by: Ya-Long Guo, Botany Institute of Botany, CAS, China
Abstract
Biodiversity is not evenly distributed among related groups, raising questions about the factors contributing to such disparities. The sunflower family (Asteraceae, >26,000 species) is among the largest and most diverse plant families, but its species diversity is concentrated in a few subfamilies, providing an opportunity to study the factors affecting biodiversity. Phylotranscriptomic analyses here of 244 transcriptomes and genomes produced a phylogeny with strong support for the monophyly of Asteraceae and the monophyly of most subfamilies and tribes. This phylogeny provides a reference for detecting changes in diversification rates and possible factors affecting Asteraceae diversity, which include global climate shifts, whole-genome duplications (WGDs), and morphological evolution. The origin of Asteraceae was estimated at ~83 Mya, with most subfamilies having diverged before the Cretaceous–Paleocene boundary. Phylotranscriptomic analyses supported the existence of 41 WGDs in Asteraceae. Changes to herbaceousness and capitulescence with multiple flower-like capitula, often with distinct florets and scaly pappus/receptacular bracts, are associated with multiple upshifts in diversification rate. WGDs might have contributed to the survival of early Asteraceae by providing new genetic materials to support morphological transitions. The resulting competitive advantage for adapting to different niches would have increased biodiversity in Asteraceae.
INTRODUCTION
Biodiversity, by its simplest definition, is the number of species within a given boundary, and is a crucial indication of ecosystem health (Loreau et al., 2001), with increasing significance for human society. Biodiversity varies dramatically among different groups of related extant organisms, including between sister lineages (Harmon, 2012; Rabosky et al., 2012). In land plants, the angiosperms (flowering plants, ~369,000 species) and gymnosperms (~1 050 species) are two sister lineages with strikingly different species numbers (Lughadha et al., 2016). Asteraceae are one of the two largest angiosperm families, comprising ~26,000 species or about 7% of extant flowering plants (Funk et al., 2009a; Christenhusz and Byng, 2016; Panero and Crozier, 2016; Wills, 2017). Asteraceae include several crops (e.g., sunflower, lettuce, artichoke), medicinal herbs, and ornamental plants, and share several floral characteristics, including a compact inflorescence (the capitulum or head) comprising one to >1,000 flowers surrounded by one or more series of involucral bracts (Leins and Erbar, 2010). The Asteraceae species richness is much greater than related families in the order Asterales, including Calyceraceae (47 spp.), Campanulaceae (2,340 spp.), Goodeniaceae (430 spp.), and Menyanthaceae (60 spp.). Asteraceae subfamilies also vary greatly in species numbers, from one species (Famatinanthoideae and Hecastocleidoideae) to approximately 17,200 species (Asteroideae). Among the other subfamilies, the next three largest are Cichorioideae (3,994 spp.), Carduoideae (2,864 spp.), and Mutisioideae (637 spp.), with the others each containing fewer than 100 species (Panero and Crozier, 2016). Asteraceae is therefore an excellent family for the analysis of the variation in biodiversity between related groups, including the possible impacts of past climate changes and morphological innovations.
Morphological and molecular evolutionary analyses require a well-resolved phylogeny. Previous analyses, largely using plastid genes, have supported the monophyly of Asteraceae (Anderberg et al., 2007), defining 45 tribes in 13 subfamilies (Funk et al., 2014; Panero et al., 2014). The relationships among the Asteraceae subfamilies and most tribes have also been resolved using plastid sequences (Fu et al., 2016; Panero and Crozier, 2016), although some uncertainties remain. In recent years, nuclear genes have been successfully used to resolve relationships among deep angiosperm lineages and within orders and families (Zeng et al., 2014, 2017; Yang et al., 2015; Xiang et al., 2016; Huang et al., 2016a; Zhang et al., 2020); for example, the relationships in Asteraceae have been analyzed using 175 or fewer nuclear genes from ~70 or fewer species (representing up to eight of the 13 subfamilies) (Mandel et al., 2017; Liu et al., 2015; Huang et al., 2016b). More recently, Mandel et al. (2019) used specific probes to capture up to 935 nuclear loci from 256 species (representing all 13 subfamilies) using largely Hyb-Seq of genomic DNAs to reconstruct the Asteraceae phylogeny in supermatrix and coalescent (ASTRAL) approaches. These results differed from the previous plastid sequence-based phylogenies in terms of the monophyly of, and relationships among, some subfamilies, with additional differences at the tribal level among the trees generated using different methods. Moreover, while the supermatrix trees supported the monophyly of Asteraceae, the coalescent tree placed a species of Calyceraceae, a previously well-supported sister family of Asteraceae, as nested within Asteraceae with low support (Mandel et al., 2019) (Figure S1). Thus, it is important to further examine Asteraceae phylogenetic relationships in additional analyses.
Whole genome duplications (WGDs) are widespread in the angiosperms and are implicated in evolutionary innovations and species diversifications (Jiao et al., 2011; Cannon et al., 2015; Li et al., 2015; Soltis et al., 2015; Yang et al., 2015; Barker et al., 2016; Xiang et al., 2016; Huang et al., 2016b). Various numbers (one to 17) of WGDs have been proposed to have occurred in Asteraceae, including one for the core Asteraceae consistently supported by genomic, phylogenomic, and Ks analyses of some or all of Asteroideae, Cichorioideae, Carduoideae, and Mutisioideae (Barker et al., 2008, 2016; Huang et al., 2016b; Badouin et al., 2017; Reyes-Chin-Wo et al., 2017; Leebens-Mack et al., 2019; Zhang et al., 2020). Another WGD event was detected for the clade encompassing the multi-tribe group called the “Heliantheae alliance” (containing Heliantheae, Eupatorieae, and other tribes) (Smith, 1975; Robinson, 1981; Yahara et al., 1989; Berry et al., 1995; Gentzbittel et al., 1995; Baldwin et al., 2002), which was also supported by analyses using Ks distribution (Barker et al., 2008) and phylogenomic methods (Huang et al., 2016b; Leebens-Mack et al., 2019). However, these previous molecular studies sampled fewer than 70 species of Asteraceae; thus, a further examination of the dates and associated taxon lineages of Asteraceae WGDs using a greater number of taxa is needed to gain insights into their possible contribution to Asteraceae biodiversity.
Here, we report the identification of 1,087 nuclear genes from the transcriptomes and genomes of 243 Asteraceae species (including 29 species overlapping with those sampled by Mandel et al. (2019)) representing all 13 subfamilies, and the reconstruction of highly supported and well-resolved Asteraceae phylogenies using both supermatrix and coalescent approaches with these nuclear genes. Our analyses allowed a comprehensive comparison of our Asteraceae phylogenies with those reported by Mandel et al. (2019). Furthermore, we present results on the divergence times of the Asteraceae lineages, shifts of diversification rates, phylotranscriptomic evidence for 41 WGDs, and the reconstruction of ancestral morphological characters. Moreover, we discuss the possible contributions of environmental changes, WGDs, and morphological innovations to the high biodiversity of Asteraceae.
RESULTS AND DISCUSSION
Asteraceae nuclear phylogeny reveals highly supported relationships among subfamilies
To reconstruct a tribal-level Asteraceae phylogeny, 243 Asteraceae species were sampled, representing all 13 subfamilies and 41 of the 45 recognized tribes (Panero and Funk, 2008; Funk et al., 2009b; Panero et al., 2014; Fu et al., 2016; Huang et al., 2016b) (149 species in Asteroideae, e.g., sunflowers, daisies, and chrysanthemums; 27 in Cichorioideae, e.g., lettuce and dandelion; 33 in Carduoideae, e.g., artichoke and thistles; and five outgroup taxa). We newly generated transcriptome and genome sequences from 121 (for one species, RNAs from two samples were sequenced) and 16 species, respectively (see Supporting Information; Tables S1, S2), plus 67 previous datasets from our lab (Zeng et al., 2014; Liu et al., 2015; Huang et al., 2016b) and 44 publicly available datasets (Tables S1, S2). To reduce possible biases caused by the use of a particular approach, we used three separate approaches to identify low-copy nuclear genes for the phylogenetic analyses (see Figure S2A for a flow chart on gene selection and Supporting Information for details).
Phylogenetic analyses using both coalescent and maximum likelihood (ML) methods with multiple datasets comprising nuclear genes yielded highly supported and consistent Asteraceae phylogenies (Figures 1, 2, S3, S14) (see Supporting Information for details). Asteraceae were monophyletic in all analyses here, forming a sister clade to Calyceraceae (three genera sampled), and the phylogeny was mostly consistent with previously reported topologies (e.g., Panero and Funk, 2008; Panero et al., 2014; Fu et al., 2016; Panero and Crozier, 2016). Asteroideae and seven other subfamilies were monophyletic with 100% support in all analyses; Famatinanthoideae and Hecastocleidoideae were monotypic; whereas Wunderlichioideae, Cichorioideae, and Carduoideae were not monophyletic (Figure 1). Seven relatively small subfamilies formed a grade of six successive sister branches of all the other Asteraceae: (i) Barnadesioideae (~92 spp.); (ii) Famatinanthoideae (one sp.); (iii) a clade with the subfamilies Mutisioideae (~640 spp.) and Stifftioideae (35 spp.), and the tribe Hyalideae (13 spp.) of Wunderlichioideae, with the topology (Mutisioideae (Stifftioideae, Hyalideae)); (iv) the tribe Wunderlichieae (34 spp.) of Wunderlichioideae (supported by 100% bootstrap support (BS) values in four trees and 98% BS in the fifth tree; Figure S15); (v) Gochnatioideae (85 spp.); and (vi) Hecastocleidoideae (one sp.).
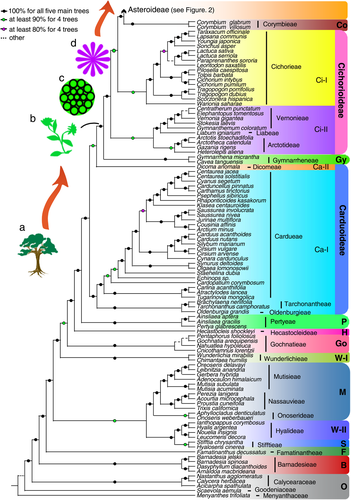
A portion of the Asteraceae phylogeny showing all subfamilies, except Asteroideae, and summaries of character reconstruction
The phylogeny is shown for all subfamilies except Asteroideae, using coalescence analyses using four gene sets (set 4: 1,087 genes; set 5: 649 genes; set 7: 384 genes; set 11: 192 genes) and maximum likelihood (ML) analysis using 192 genes (set 11) obtained as explained in Figure S2 (see Supporting Information for details). The individual phylogenies are shown in Figures S3–S14, with support values shown in Figure S15. To the right of generic/specific names are tribe names, with subfamily names abbreviated as follows: B, Barnadesioideae; F, Famatinanthoideae; S, Stifftioideae; W-II, Wunderlichioideae-II; M, Mutisioideae; W-I, Wunderlichioideae-I; Go, Gochnatioideae; H, Hecastocleidoideae; P, Pertyoideae; Gy, Gymnarrhenoideae; Co, Corymbioideae. The subfamilies Cichorioideae and Carduoideae are paraphyletic and are indicated with two vertical bars, with the clades labeled as Ci-I/Ci-II and Ca-I/Ca-II, respectively. The change of habit from woody (a) to herbaceous (b) according to ancestral character reconstruction (Figure S36) is estimated to have occurred at the root node of Gymnnarrhenoideae – Asteroidaee. The change of capitulum type is according to the analysis shown in Figure S39 and shown here from an ancestral and basal discoid with only disc florets (c) to ligulate capitulum with only ligulate florets (d) in the subfamily Cichorioideae and radiate capitulum (e in Figure 2) characterized by having both ray and disc florets and found in most members of Asteroideae and some in Cichorioideae II.
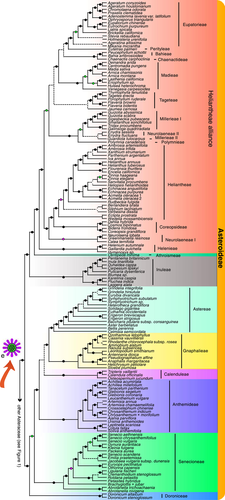
A portion of the Asteraceae phylogeny with subfamily Asteroideae and summaries of character reconstruction
The phylogeny shown here is for the largest subfamily Asteroideae, with 20 of its 21 tribes (except Feddeeae), summarized from results as described in the legend of Figure 1. The morphological change from discoid (c in Figure 1) to radiate (e) with both ray and disc florets, according to the ancestral character reconstruction is detailed in Figure S39. The latter type is found in most members of Asteroideae, with an important exception being members of the tribe Eupatorieae, which likely lost the ray florets following the separation from the small tribe Perityleae.
Mandel et al. (2019) reported that Barnadesioideae were sister to the other Asteraceae species in the supermatrix trees only, whereas their relationships with Calyceraceae and the remaining Asteraceae were poorly resolved in the coalescent tree (Figure S1). Furthermore, the phylogenetic relationships of Famatinanthoideae, Mutisioideae, Stifftioideae, and Hyalideae in the two supermatrix trees presented by Mandel et al. (2019) were the same as in our phylogeny; however, in their coalescent tree, Famatinanthoideae were sister to a clade comprising Stifftioideae and Hyalideae (97% BS), with Mutisioideae being the next sister group separating them from the remaining Asteraceae (71% BS). Moreover, Wunderlichieae, Gochnatioideae, and Cyclolepis (not sampled here) formed a clade (64% BS/1.0 Bayesian posterior probability (PP) in the supermatrix trees; and 100% BS in the coalescent tree), although the placement of Cyclolepis was inconsistent (Figure S1). Further studies are therefore needed to clarify the phylogenetic positions of Wunderlichieae, Gochnatieae, and Cyclolepis.
Several clades, including Hyalideae, Mutisioideae, and Stifftieae, were highly supported in all our analyses as well as in the supermatrix analyses presented by Mandel et al. (2019). Previously, Hyalideae were found to be sister to Wunderlichieae and were thus placed in Wunderlichioideae (s.l.) (52/71/84% BS; 0.91/0.998/0.99 PP) in analyses performed using 10–14 plastid loci (Panero and Funk, 2008; Panero et al., 2014; Panero and Crozier, 2016). By contrast, analyses using nuclear ITS sequences (91% BS/1.0 PP; Funk et al. (2014)) and numerous protein-coding nuclear genes (here and also by Mandel et al. (2019)) all highly supported the sister relationship of Hyalideae and Stifftieae.
Among the remaining Asteraceae, Pertyoideae (~80 spp.) are sister to an extremely large clade (>24,000 spp.) that includes the three largest subfamilies, Asteroideae, Cichorioideae, and Carduoideae, as well as two very small subfamilies, Corymbioideae and Gymnarrhenoideae (Figures 1, 2). This position of Pertyoideae is also supported by recent analyses using ITS data (Funk et al., 2014) and hundreds of nuclear sequences (Funk et al., 2014; Mandel et al., 2019) (Figure S1). However, in plastid-based phylogenies, Pertyoideae are sister to the clade Gymnarrhenoideae–Asteroideae (Panero and Funk, 2008; Panero et al., 2014; Panero and Crozier, 2016). Pertyoideae are the only subfamily in which distribution is restricted to East Asia, and that produce corollas with five irregularly split lobes intermediate between the typical ligulate (such as those of Cichorieae) and tubular flowers (such as those in Cardueae and some Asteroideae). In addition, the basal chromosome numbers in Pertyoideae are x = 12–15 (Wang, 2009; Zhang, 2013), unlike the presumed ancestral Asteraceae chromosome number of nine (Semple and Watanabe, 2009). Nevertheless, some morphological features of Pertyoideae, such as short style branches with papillae on the abaxial surface and relatively simple pollen surfaces, resemble those of the early-divergent branches of the Asteraceae rather than Carduoideae (Katinas et al., 2008). Among the single-gene family trees of the 192 genes in set 11 (Figure S2), the position of Pertyoideae reported here was supported by 52 trees with BS values of more than 75%, and by 97 additional trees with BS values between 50% and 75%, but no trees supported the previously published placement based on the analyses of plastid genes (e.g., Panero and Funk, 2008; Panero et al., 2014; Fu et al., 2016; Panero and Crozier, 2016). Future studies with expanded sampling could improve our understanding of the placement of Pertyoideae.
The mostly Old World subfamilies Carduoideae and Cichorioideae, as previously defined (Funk et al., 2014; Panero et al., 2014; Fu et al., 2016), were paraphyletic in all of our phylogenetic analyses. All four tribes in Carduoideae formed two clades, which are indicated, respectively, as Carduoideae I and II (Figure S15, and abbreviated, respectively, as Ca-I and Ca-II in Figures 1, S1). Carduoideae I includes three tribes, Cardueae, Oldenburgieae, and Tarchonantheae, with maximum support, whereas Carduoideae II contains the fourth small tribe, Dicomeae, which is consistently placed as sister (BS values of 100%, 90%, 89%, 87%, and 87%; Figure S15) to the clade Gymnarrhenoideae–Asteroideae. Carduoideae I (Figures 1, S1, S15) is the sister lineage to the clade Carduoideae II–Asteroideae. The coalescent (ASTRAL) tree presented by Mandel et al. (2019) grouped all four tribes of Carduoideae in a single clade, whereas the supermatrix ML analyses placed the combined Tarchonantheae and Oldenburgieae clade, the Dicomeae clade, and the Cardueae clade as three separate branches in a grade of successive sisters to the clade of Gymnarrhenoideae–Asteroideae (Figure S1). On the other hand, phylogenies using plastid genes strongly supported the inclusion of Dicomeae in the Carduoideae (Fu et al., 2016; Panero and Crozier, 2016). Gene flow among Dicomeae, Cardueae, Tarchonantheae, and Oldenburgieae and the underlying assumptions of the different methods used are possible explanations for the different relationships seen in these phylogenies.
The circumscription of Cichorioideae has changed several times, even very broadly, from containing only one tribe, Cichorieae, to encompassing all Asteraceae members other than Asteroideae (see review by Funk and Chan, 2009). The recent definition of the Cichorioideae was based on analyses using plastid sequences (Funk et al., 2014; Panero et al., 2014; Fu et al., 2016), but it lacks the support of morphological synapomorphy. Among the four Cichorioideae tribes sampled here, Cichorieae (Figures 1, S1, S15) were sister to the clade Asteroideae + Corymbioideae (BS = 89–100% in the four phylogenies) (Figure S15). On the other hand, three other tribes, Vernonieae, Liabeae, and Arctotideae, form a clade (BS of 100%, 100%, 99%, and 93%) sister to the clade Asteroideae + Corymbioideae + Cichorieae (Figure 1). The paraphyly of Cichorioideae was also supported by Mandel et al. (2019) (Figure S1). Thus, Cichorieae are proposed as Cichorioideae s.s. (indicated as Cichorioideae I in Figure S15, and abbreviated as Ci-I in Figures 1, S1), whereas the other three tribes, Vernonieae, Liabeae, and Arctotideae, are proposed to form a separate subfamily (indicated as Cichorioideae II in Figure S15 and abbreviated as Ci-II in Figures 1, S1). The difference in the position of Cichorieae relative to the other tribes between the phylogenies using nuclear or chloroplast genes could be explained by a possible hybridization in the history of Cichorieae.
Resolution of the relationships among the Asteraceae tribes
In addition to the relationships among Asteraceae subfamilies, the nuclear phylogenies here also provide strong support for the relationships among tribes (Figures 1, 2, S3–S15). Among the 41 tribes in this study, 30 were monophyletic with 100% support (Figures 1, 2), but in the subfamily Asteroideae, Millerieae and Neurolaeneae were not monophyletic (Figure 2). The remaining nine tribes were represented by one species each (two corresponding to monotypic subfamilies). In addition to the relationships among subfamilies of Asteraceae, the nuclear phylogenies generated here also provide strong support for the relationships among tribes (Figures 1, 2, S3–S15). The vast majority of the tribes represented in this study were also sampled in Mandel et al. (2019) (Figure S1), except Polymnieae (Polymnia) in Asteroideae (Figures 2, S1). The sister relationship of Mutisieae and Nassauvieae, and that of Oldenburgieae and Tarchonantheae, were both also supported by phylogenies developed using plastid genes (Panero and Funk, 2008; Panero et al., 2014) and nuclear genes (Mandel et al., 2019). Within Cichorioideae II, the tribes Vernonieae and Liabeae are sisters, but more distant to Arctotideae (Figure 1), consistent with their positions in previous phylogenetic analyses (Figure S1). In Arctotideae, Heterolepis (Arctotideae III) is sister to Arctotideae I (Arctotidinae) in our analyses, but sister to Arctotideae II (Gorteriinae) in the analyses performed by Mandel et al. (2019) (Figure S1), and it has varying positions in previous analyses performed using different datasets and different outgroups (Funk et al., 2004, 2009a). Future analyses with other methods and more taxon sampling may shed light on the relationships among Heterolepis and other Arctotideae species.
The subfamily Asteroideae were previously divided into three supertribes: Asterodae, containing four tribes; Helianthodae, containing 15 tribes; and Senecionodae, with only one tribe, Senecioneae (Robinson, 2004). The analyses performed here (Figures 2, S15) support the monophyly of Asterodae with 100% BS. Among the four Asterodae tribes, Astereae and Gnaphalieae were sister clades (100% BS), with Anthemideae and Calenduleae splitting successively and as sister to the other Asterodae. Although the sister relationship between Astereae and Gnaphalieae was also recovered in nuclear phylogenies performed by Liu et al. (2015) using 49 nuclear genes and by Mandel et al. (2019), Anthemideae were placed differently in previous analyses, either as sister to Astereae with high support in plastid phylogenies (Panero and Funk, 2008; Panero et al., 2014; Panero and Crozier, 2016), or as sister to Senecioneae in nuclear phylogenies with limited samplings by Liu et al. (2015) and from the supermatrix analyses with 75% BS in Mandel et al. (2019) (Figure S1). The previously reported sister relationship between Anthemideae and Senecioneae suggested that Asterodae might not be monophyletic. This, in combination with Astereae and Anthemideae being sisters in the plastid phylogenies, led Liu et al. (2015) to propose a hybrid origin of Anthemideae from a cross between parental lineages related to Astereae and Senecioneae, respectively, which was also supported by some morphological characters (see discussion by Liu et al., 2015). By contrast, the sister relationship of Anthemideae and Senecioneae was supported by nuclear gene analyses of three species in each tribe (Liu et al., 2015), and in 12 and 16 species in Anthemideae and Senecioneae, respectively (Mandel et al., 2019). However, this reported phylogenetic relationship did not include the Senecioneae genus Abrotanella, which have disciform capitula with four-lobed corollas, unlike most Senecioneae, which have discoid or radiate capitula with five- and/or three-lobed corollas (Nordenstam, 2007). The sampling here included 16 Anthemideae species in 12 genera and 18 Senecioneae species in 14 genera, including two Abrotanella species. Our findings placed Abrotanella as sister to the other Senecioneae with maximal support (Figure 2). The maximally supported monophyly of Asterodae (including Anthemideae) and Senecioneae (including Abrotanella) suggested that the increased sampling or inclusion of Abrotanella here was important for achieving the highly supported resolution of these groups, and does not support Senecioneae as a possible parental lineage of Anthemideae.
In addition, the genus Doronicum was resolved here as sister to the combined clade of Asterodae and other members of Senecioneae (Figures 2, S15), supporting its designation as the tribe Doroniceae proposed by Panero (2005) and adopted by Fu et al. (2016) in their systematic arrangements for Asteraceae from China. Doronicum was not sampled by Mandel et al. (2019). Doronicum was traditionally placed in Senecioneae, primarily according to gross morphology; however, its relationship with members of the Senecioneae was poorly resolved in the ITS- and plastid-based phylogenies (Pelser et al., 2010; Fu et al., 2016). Consequently, Doronicum was not assigned to a previously defined Senecioneae subtribe (Nordenstam et al., 2009). The strongly supported position of Doronicum here clearly separates it from Senecioneae and argues for its designation as a distinct tribe. Both Doronicum and Abrotanella were members of Senecionodae (Nordenstam, 2007), but the phylogeny generated here suggests that Senecionodae are not monophyletic.
The supertribe Helianthodae contains 15 tribes, all sampled here except for the monotypic Feddeeae (Figure 2). The relationships we identified among the Helianthodae tribes are consistent with those reported by Mandel et al. (2019), except for some taxa that were only sampled here, including one tribe (Polymnieae, containing Polymnia) and three genera (Enydra, Guardiola, and Jaumea, in the tribes Neurolaeneae, Millerieae, and Tageteae, respectively). All phylogenies herein provide strong support for the monophyly of the supertribe and seven of the 10 tribes containing two or more species: Bahieae, Coreopsideae, Eupatorieae, Helenieae, Heliantheae, Inuleae, and Madieae. In contrast, Tageteae were monophyletic only in the ML analysis of 192 genes (set 11); Jaumea did not group with other Tageteae genera in the coalescent phylogenies (Figures S3–S15), similar to results of the analysis using plastid sequences (Fu et al., 2016). Moreover, Millerieae and Neurolaeneae were not monophyletic; the genus Enydra (two species sampled here; previously in Neurolaeneae (Panero, 2007)) was nested within the Millerieae, which were represented here by six genera. In a previous plastid phylogeny (Fu et al., 2016), Enydra was sister to a clade of three other Neurolaeneae genera (also sampled here), but the grouping of Enydra with the other Neurolaeneae was not strongly supported (<50% BS/0.95 PP). Millerieae and Enydra are both pantropical, whereas the other Neurolaeneae are all restricted to the Americas (Panero, 2007). Thus, the phylogenetic position of Enydra among Millerieae genera and their geographical distribution may support a proposed expansion of Millerieae to include Enydra.
Among the Helianthodae tribes, Inuleae and Athroismeae were successively diverged from the other tribes with maximum support, and the remaining tribes formed a strongly supported clade, referred to as the “Heliantheae alliance” (Panero, 2007; Baldwin, 2009) (Figures 2, S15). Within the Heliantheae alliance, Helenieae were the first to diverge, with the other tribes forming three major clades. The first major clade includes Neurolaeneae (excluding Enydra), Heliantheae, and Coreopsideae. Members of these groups produce head inflorescences with bracts (referred to as paleae or receptacular bracts) subtending the florets/achenes. Heliantheae and Coreopsideae were strongly supported sisters (Figure 2), which was also supported by previous analyses using ITS data (Baldwin et al., 2002) or dozens of nuclear genes (Liu et al., 2015); however, different topologies were recovered using plastid sequences (Jansen et al., 1991; Panero and Funk, 2002; Panero et al., 2014). The inconsistent positions of Coreopsideae between the nuclear and plastid phylogenies were potentially due to hybridization events (Panero, 2007; Liu et al., 2015), which might also be true for Heliantheae. In the second clade we identified with six tribes, Eupatorieae and Perityleae were strongly supported as sisters (Figure 2), as were Bahieae and Chaenactideae, in agreement with previous studies (Panero and Funk, 2002; Panero and Crozier, 2016). Tageteae were sister to Madieae and the weakly supported clade ((Chaenactideae, Bahieae), (Perityleae, Eupatorieae)). These six tribes are mostly epaleate (Panero, 2007).
The third major clade was sister to the second clade and contains Millerieae + Enydra + Polymnieae, the latter of which contains only one genus, Polymnia, with three species distributed in eastern North America. The Neurolaeneae genus Enydra is pantropical, whereas Millerieae genus Guardiola is mostly found in Mexico and is aquatic (Figure 2). Polymnia is superficially similar to other genera in Millerieae but was placed in the Heliantheae subtribe Polymniinae by Robinson (1978), who considered most species previously placed in Polymnia to be members of the genus Smallanthus (Millerieae). Polymnieae were once placed in Millerieae as the subtribe Polymniinae (Robinson, 1978), but in the plastid phylogenies they constituted a distinct clade separate from other Millerieae species (Panero and Funk, 2002; Panero, 2007). Moreover, most members of Millerieae were closely related to Heliantheae in the plastid phylogenies, but Enydra was included in the Neurolaeneae clade (Panero, 2007; Panero and Funk, 2008); these differences in nuclear and plastid phylogenies thus suggest a possible hybrid origin.
Asteraceae origin was estimated in the Late Cretaceous, while most tribes diverged before the Oligocene
Environmental factors play major roles in shaping biodiversity. To obtain clues about possible historical environmental influences on biodiversity in Asteraceae, we estimated the times of the origins of, and divergences among, the Asteraceae lineages using the newly reconstructed nuclear phylogeny (Figures 1, 2) and the 192 nuclear genes (set 11 in Figure S2). We performed molecular clock analyses using calibrations with 15 fossil constraints, including seven Asteraceae fossils (Table S2), and a secondary calibration for the crown node of the Eudicots (Zanne et al., 2014; Tank et al., 2015). A fossil pollen, Tubulifloridites lilliei type A (referred as Tl-typeA hereafter) was reported as being related to early members of Asteraceae, but with uncertain placement (Barreda et al., 2015). To test the effects of the use and position of Tl-typeA on age estimation, it was either not included (calibration set 1) or included with differing placements (sets 2–5) (see Supporting Information for details). The results from the r8s and BEAST analyses were nearly identical among calibration set 1 and sets 3–5 (Figures S16–S25); thus, only the results from using calibration sets 1 and 2 are discussed below.
The mean ages were similar along the backbone from the r8s and BEAST analyses using the same calibration set (Figure 3; Table S3). Particularly, the estimated ages of the most recent common ancestors (MRCAs) of Asteraceae, Calyceraceae, Goodeniaceae to Menyanthaceae, and of Heliantheae alliance varied by no more than 3 My between the two methods. Variations of 10–15 My were found in the ages of some lineages, such as Cichorioideae I, Carduoideae I, and Cichorioideae II. On the other hand, estimations with the fossil calibration set 1 (without the pollen fossil) and set 2 (with the pollen fossil at the position assigned by Barreda et al., 2015) performed using the same method differed by fewer than 10 My for most nodes, except those of the crown Barnadesioideae and one of its two subclades (the MRCA of Dasyphyllum and Arnaldoa) (Figures S26, S27). Thus, the results from both methods and each fossil calibration set all suggest the origin of Asteraceae in the middle of the Late Cretaceous, with the separation of the subfamilies before or near the Cretaceous–Paleocene boundary at ~66 Mya, and divergence of most of the tribes before the Oligocene, or during the Eocene or Paleocene (Figure 3). Gymnarrheneae, Cardueae, Pertyeae, Hecastocleideae, Gochnatieae, Wunderlichieae, Famatinantheae, and Barnadesieae probably arose before the end of the Cretaceous (Figure 3).
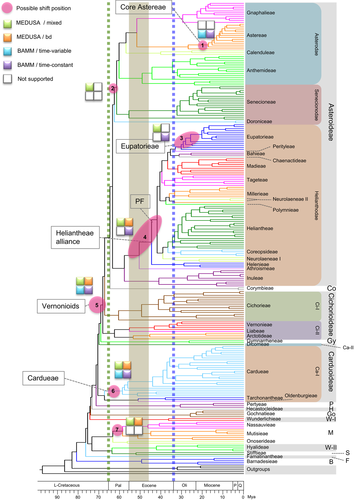
Molecular clock estimates of divergence times and diversification rate shifts in Asteraceae
This figure presents the subtree containing members of Asterales retrieved from the result of age estimation with calibration set 1. Green and blue dotted lines indicate the boundaries of Cretaceous-Paleocene and Eocene-Oligocene, respectively. The brown stripe corresponds to the hottest period of the Cenozoic era. Multiple specific molecular clock analyses are shown in Figures S16–S27. The positions for increases of net diversification rate resulting from individual analyses are collectively depicted with colored circles. Colored blocks indicate the analysis supporting a shift at the indicated position. Detailed results from each analysis are shown in Figures S28–S34.
Including Tl-typeA on the Dasyphyllum stem led to an estimated age of the crown Asteraceae similar to those reported by Barreda et al. (2015); however, this result (set 2) differed substantially from those with different placements of Tl-typeA (sets 3–5). The estimated ages of the crown Asteraceae from the latter three placements were similar to each other and to the age estimated without the inclusion of Tl-typeA, indicating an obvious impact of assigning Tl-typeA to Dasyphyllum. Because this placement is controversial (Panero, 2016), we accept the more conservative and consistent results from the other four estimations and discuss the ages from calibration set 1 below. Using calibration set 1, Asteraceae and its sister clade, Calyceraceae, were estimated to have diverged ~83 Mya. The Barnadesioideae then separated from the other Asteraceae subfamilies ~81 Mya, with seven other subfamilies progressively diverging during the next 10 My. Throughout the late Cretaceous when these subfamilies separated, the climate was much warmer and more humid than the present (Linnert et al., 2014), suggesting that higher temperatures might have promoted early Asteraceae diversification (Davies et al., 2004; Jablonski et al., 2006; Jansson and Davies, 2008).
Except for Stifftioideae, all other subfamilies, including the three largest subfamilies (Asteroideae, Cichorioideae, and Carduoideae, comprising 69%, 16%, and 11% of Asteraceae species, respectively) (Panero and Crozier, 2016), diverged before the Cretaceous–Paleocene boundary when massive extinctions occurred (at 66 Mya) (Figure 3) (Jablonski and Chaloner, 1994). Also, the supertribes Asterodae and Helianthodae and the tribe Senecioneae (Figure 3) separated ~62–61 Mya, with further divergences of most of the ~20 Asteroideae tribes in the Eocene. Further divergences among some tribes/subtribes, and also genera, occurred after the Eocene–Oligocene boundary alongside great climate changes and numerous extinctions (Ivany et al., 2000). In addition to the possible roles of climate factors, the massive extinctions at the Cretaceous–Paleocene and Eocene–Oligocene boundaries, which likely freed numerous ecological niches, could therefore also have facilitated the diversification of the largest subfamily, Asteroideae.
The ages estimated here are generally older than those reported in previous studies, including the estimated age of the Asteraceae stem of ~50 My reported in studies with sampling at higher (e.g., across several families or orders) or lower (e.g., of a tribe) taxonomic levels (Bremer et al., 2004; Barres et al., 2013; Beaulieu et al., 2013; Jabaily et al., 2014; Magallón et al., 2015; Park and Potter, 2015; Tank et al., 2015). The ages estimated here using calibration set 1 (Figures 3, S16, S17) are also older than those reported by Panero and Crozier (2016) using 11 plastid genes and one noncoding region (85 species in 39 tribes), such as the age estimated here for the crown Asteraceae of ~81 My compared with the ~65 My reported by Panero and Crozier (2016). However, our estimated age for the crown Asteraceae is close to that (83 Mya) reported by Mandel et al. (2019). The differences in estimated ages might be due to the sequence datasets (nuclear vs. plastid genes), gene numbers, and/or taxon sampling, as well as the calibrations (Table S2).
Multiple increases in the diversification rate in the Asteraceae
Changes in species richness could be due to either increases or decreases in diversity, which could be estimated by the analysis of shifts in the rate of diversification using a reference phylogeny. To better understand the history of diversity changes in Asteraceae, we estimated the diversification rates and identified the approximate positions of rate shifts during Asteraceae evolution using the MEDUSA (Alfaro et al., 2009) and BAMM methods (Rabosky, 2014; Rabosky et al., 2014b; Shi and Rabosky, 2015). The positions with potential major accelerations in net diversification rate obtained using both methods (each with two different models) are collectively illustrated in Figure 3. For the analyses using MEDUSA (with mixed and birth–death models; Figures S28, S29), we collapsed the tree tips to the tribal level and used the species number of each tribe as the species richness. The result showed six accelerations in net diversification rates (red circles) and one deceleration (blue circle) (Figure S28). Among the upshifts, four with 2.7-, four-, three-, and twofold increases relative to the background (circles 2–5), respectively, are associated with the Vernonioid clade (Asteroideae–Cichorioideae II), the Heliantheae alliance excluding Helenieae (also called the phytomelanic (dark-colored) fruit (PF) clade), the tribe Eupatorieae, and the clade comprising two supertribes (Asterodae and Senecionodae). Rate accelerations were also found at nodes leading to the Nassauvieae + Mutisieae clade and the tribe Cardueae, resulting in approximately twofold increases in the diversification rate.
We also used BAMM with the complete tree in Figure 3 and the sampling fraction data (Table S4), performing separate analyses using both time-variable and time-constant algorithms. Both algorithms produced the same best shift configurations (the one with maximum a posteriori probability; MAP configuration) with three rate shifts (Figure S30): at the Vernonioid clade (circle 5), the Cardueae clade (circle 6), and the core Astereae clade (circle 1). These shifts can also be observed in the rate through time plots (Figure S30). The shifts at the Heliantheae alliance/PF (circle 4) and the Eupatorieae clade (circle 3) were also supported by BAMM under the time-constant algorithm in a further investigation of the results (Figures S32–S34; Table S10).
In summary, all four analyses performed here strongly support upshifts of the net diversification rates at the Vernonioid clade (circle 5) and at tribe Cardueae (circle 6) (Figure 3), affecting the largest subfamilies, Asteroideae, Cichorioideae II (Vernonioideae), and Carduoideae. The next likely position for a diversification rate upshift is among the nodes from the PF clade to the Heliantheae alliance, and may even include Athroismeae (circle 4); finding an event here possibly benefited from a greater sampling of this tribe. This group is positioned within the large supertribe Helianthodae and includes many tribes that further expanded after the Eocene–Oligocene boundary during the dramatic climate changes and mass extinctions. Another possible upshift took place near the divergence of Eupatorieae (circle 3), which expanded greatly after the Eocene–Oligocene boundary. Mutisieae (or the Mutisieae + Nassauvieae clade) (circle 7) is another group with a possible rate increase that expanded after the Cretaceous–Paleocene boundary. Similarly, the clade Asterodae + Senecioneae and the core Astereae also had possible diversification rate increases; the earlier divergence among the tribes occurred after the Cretaceous–Paleocene boundary, whereas the later divergence within Astereae took place after the Eocene–Oligocene boundary. Among these, the upshifts at the PF and Vernonioid clades (circles 4 and 5) were also reported by Panero and Crozier (2016), and those marked with circles 2 and 4 were consistent with those proposed by Mandel et al. (2019).
Detection of multiple WGD events
Several WGDs have previously been detected in Asteraceae using genomic, phylogenomic, and Ks analyses of 70 or fewer species (Barker et al., 2008, 2016; Huang et al., 2016b; Badouin et al., 2017; Reyes-Chin-Wo et al., 2017; Leebens-Mack et al., 2019; Zhang et al., 2020). The newly resolved Asteraceae phylogeny generated here from large-scale datasets of 243 species representing all subfamilies and almost every tribe provide an unprecedented opportunity to detect Asteraceae WGDs and place them phylogenetically. We investigated WGD by reconstructing trees of 5 282 orthologous groups (OGs) and comparing them with the reference phylogeny, detecting numerous clusters of gene duplications (GDs) as evidence for a WGD at one of multiple nodes of the Asteraceae phylogeny (see Materials and Methods). According to the strength of the GD evidence, we propose nine WGDs and 32 candidate WGDs (Figures 4, S35), including WGD1 shared by Calyceraceae and Asteraceae, WGD2 shared by the core Asteraceae (Asteroideae–Mutisioideae/Stifftioideae), and WGD3/WGD4 at successive nodes shared by tribes of the Heliantheae alliance (without/with Helenieae, respectively).
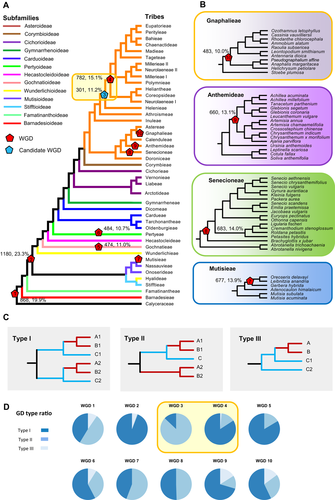
A summary of whole-genome duplications (WGDs) detected in Asteraceae
(A) The lineages in each of 13 subfamilies are represented by colored lines. Nine detected WGDs are marked as red pentagons, with one candidate WGD (WGD3) marked as a blue pentagon, with the detected GD numbers and percentages. (B) The node of WGDs within four tribes are marked. (C) Three types of the topologies of retained duplicates are illustrated. For the two sub-clades of taxa derived from one node, Type I has retention of both duplicates in both subclades. Type II lacks one copy in the small sub-clade (blue), and Type III lacks one copy in the large sub-clade (red). (D) The ratio of three types of the nine WGDs and the candidate WGD (WGD3). Additional information for WGD events is shown in Figure S35.
WGD1 and WGD2 have also been detected in previous studies (Barker et al., 2016; Huang et al., 2016b), and are consistent with those reported in analyses including multiple angiosperm families and small numbers of Asteraceae species (the XASTβ event described by Leebens-Mack et al., 2019), and WGD #23 described by Zhang et al. (2020)). It is worth noting that, following WGD2, the chromosome base number decreased from 27 to 10, and the species dispersed from South America to Africa/Asia–Eurasia (Funk and Chan, 2009; Semple and Watanabe, 2009). After WGD3, the chromosome base number changed once again from 10 to 19, and the species dispersed from Africa/Asia–Eurasia to North America (Funk and Chan, 2009; Semple and Watanabe, 2009). However, in our analysis, WGD3 shows much fewer GDs than WGD4 (301 vs. 782, respectively), and most of the GDs are of the Type II pattern (~87%), indicating that only one gene copy was detected in Helenieae species in most OGs (Figure 4C, 4D). WGD4 (without Helenieae) is also consistent with the XASTα event described by Leebens-Mack et al. (2019). These results have three possible explanations: (i) the WGD event occurred at the Heliantheae alliance (WGD3), but both copies were lost in Helenieae for most OGs; (ii) the WGD event occurred at WGD3, but the variable substitution rates among the Helenieae and other lineages of the Heliantheae alliance caused many of the gene duplications to apparently support WGD4; or (iii) there was a hybridization event between Helenieae and the ancestor of the other Heliantheae alliance species shortly after their divergence.
There are also large numbers of GDs at the crown node of the subfamilies Gochnatioideae (WGD5) and Pertyoideae (WGD6); and within the tribes Mutisieae (WGD7), Senecioneae (WGD8), Anthemideae (WGD9), and Gnaphalieae (WGD10) (Figure 4B). We also detected 31 other clusters of GDs, providing evidence for candidate WGDs (Supporting Information text; Figure S35). In summary, previous studies using a relatively small number of species (Barker et al., 2016; Huang et al., 2016b; Leebens-Mack et al., 2019) and the present analysis with much greater sampling reached the same conclusion about WGDs at nodes shared by many lineages (WGD1, WGD2, and WGD3/WGD4). In addition, other tribes have experienced independent WGD events, affecting groups with very high species richness (>1 000 species). Of the 32 candidate WGDs (Figure S35), a large majority were detected in the largest subfamilies, including 19 events in Asteroideae, three in Cichorioideae (including the tribe Cichorieae), and two in Carduoideae. Nevertheless, some WGDs were associated with small subfamilies or tribes, such as Gochnatioideae (~70 spp.), Pertyoideae (~80 spp.), and Mutisieae (~254 spp.), suggesting that a WGD alone might not be sufficient for increased diversity and that other factors, such as environmental conditions, are also important. This is consistent with a previous analysis of multiple WGDs throughout the angiosperms (Ren et al., 2018).
Ancestral states of morphological characters
Morphological innovations can afford evolutionary advantages and promote divergence and biodiversity; therefore, we examined the morphological evolution of Asteraceae in the context of the nuclear phylogeny presented here with the aim of identifying a link between morphological innovation and organismal diversity. We traced the ancestral states and histories of seven evolutionarily significant characters, including the habit, pappus, and five floral traits (Figures S36–S42). The Asteraceae ancestor was most likely woody, with epaleate receptacles, a solitary homogamous capitula with isomorphic and discoid florets, and a capillary/plumose pappus, which is largely consistent with the estimations of Bremer (1994) and Panero et al. (2014). One important morphological change along the backbone is from the ancestral woody habit to the herbaceous habit (Figures 1, S36) at the last common ancestor of multiple subfamilies, including the Gymnarrhenoideae and Asteroideae, with a likelihood value 0.948. This estimated change in habit is older than the root node of the Cichorioideae–Asteroideae (0.899) estimated by Panero et al. (2014). It is possible that the transition to herbaceousness could have occurred later than the estimate here, as our sampling did not include some of the woody species in Cichorioideae II (Karis et al., 2009; Robinson, 2009; Robinson and Funk, 2009). Regardless of the precise position of the transition to the herbaceous habit, the woody habit in early Asteraceae history is supported by the woodiness of members of the subfamilies Barnadesioideae, Famatinanthoideae, Stifftioideae, Wunderlichioideae, Gochnatioideae, Hecastocleidoideae, and Pertyoideae in a grade of early divergent sister lineages of most of Asteraceae. This is further supported by the presence of woody members in the tribes Onoserideae and Nassauvieae, as successive sisters of other Mutisioideae, and in the tribes Oldenburgieae and Tarchonantheae that form a sister clade to the Cardueae (Figure S36). On the other hand, most members of the large subfamilies Asteroideae and Cichorioideae (s.l.) and the tribe Cardueae are herbaceous, although habit transitions have also occurred later in Asteraceae history, even among closely related species (e.g., Panero et al., 1999), with woody members in these groups likely derived secondarily from herbs.
Asteraceae are characterized by a head inflorescence (capitulum) with sessile florets surrounded by bract-like organs in a compact structure (Funk et al., 2009b), which can be solitary or part of a higher order inflorescence (capitulescence) (Figure S37). The florets in a capitulum can be uniformly bisexual (homogamous) or exhibit sexual differentiation between the outermost and inner florets (heterogamous) (Figure S38). In addition, the corolla of florets exhibits several morphologies, including the actinomorphic (radially symmetric) disc florets found in several subfamilies, the zygomorphic (bilaterally symmetric) ligulate florets of Cichorieae, and the zygomorphic ray florets on the periphery of heads exemplified by members of Asteroideae. Thus, discoid heads contain only disc florets, radiant heads have marginal disc florets with an enlarged corolla, while ligulate heads contain only ligulate florets, which have a corolla with a five-lobed outer lip. Radiate capitula are found in sunflowers (Helianthus) and most Asteroideae, and comprise outer pistillate or neutral ray florets and inner bisexual disc florets. On the other hand, in disciform heads, the outer florets are pistillate but lack the large corolla of ray florets.
The ancestral character analysis here supports an inflorescence transition from solitary to capitulescence prior to the divergence of the clade Hecastocleidoideae (0.983) (Figure S37), similar to the estimation of Panero et al. (2014). The homogamous and discoid capitulum were estimated as the ancestral state at the Asteraceae root node (with 99.36% and 99.95% likelihood values, respectively) (Figures S38, S39). Previously the ancestral discoid capitulum had a likelihood value of only 48% (Panero et al., 2014), probably because of differences in the coding of capitulum type for some taxa in Mutisioideae and Hyalideae. In these groups, the outer florets have zygomorphic corollas with a tri-lobed outer lip and a much smaller bi-lobed inner lip. Thus, these capitula are referred to as radiate-like and are different from the true radiate capitula, which have no inner lip in the corolla of outer florets. The genes contributing to the formation of radiate-like capitula and radiate capitula are also different (Chen et al., 2018). In most Asteroideae, heterogamous and radiate capitula are the symplesiomorphies; the morphological and sexual differentiations between outer ray florets and inner disc ones make the capitulum function as a single larger flower. These large “flower-like” heads are again often arranged in a group or series. Such higher order inflorescences (capitulescences) have been recognized as having great success in attracting pollinators (Stuessy et al., 1986; Celep et al., 2014), thereby contributing to the diversification of this largest Asteraceae subfamily comprising more than 15 000 species. Radiate capitula were also found to have arisen independently in Arctotideae, Liabeae, and Oldenburgieae. On the other hand, the transition of radiate to discoid or disciform capitula, the outer florets of which lack the large outer lip of rays, was estimated to have occurred independently several times, with one affecting the entire Eupatorieae tribe.
In most Asteraceae, the floret calyx persists after anthesis and is called the pappus. The pappus remains attached to the inferior fruit (achene) during fruit development and even after maturity. It has different morphological types, such as capillary (hair-like), plumose (feather-like), and scaly. Some Asteraceae also have a bract-like organ (called the palea, or receptacular bract) subtending all or some florets on a receptacle. Both the pappus and the palea serve to protect the developing fruit, and the pappus often facilitates the dispersal of achenes, such as by wind for dandelion and many others (Stuessy and Spooner, 1988; Stuessy and Garver, 1996). Some Asteraceae members lack the pappus (and are therefore epappose); these taxa are found mostly in the Anthemideae and partly in the Heliantheae alliance, and generally possess receptacular bracts (Stuessy and Garver, 1996). In the present study, the pappus was estimated to be capillary (64.7%) or plumose (30.0%) for the root node of Asteraceae. More specifically, the capillary pappus was supported to be the ancestral state for most nodes except the earliest divergent clade containing Barnadesioideae and the Heliantheae alliance. This is different from the Asteraceae ancestral state of a scaly pappus with a defensive function proposed by Stuessy and Garver (1996). The most common pappus type in Barnadesioideae is the plumose pappus (Stuessy et al., 2009), which likely facilitates seed dispersal over a greater distance. Considering the origin and early evolution of Asteraceae in much warmer climates and more closed habitats (e.g., forest) than the conditions today, the dispersal function of a plumose/capillary pappus would be more important in the early evolution of Asteraceae. On the other hand, the defense function of a scaly pappus would be more important against herbivores, especially for many species of the Heliantheae alliance with larger achenes. For the Heliantheae alliance, various kinds of scaly pappus and paleate receptacles for the protection and dispersal of achenes might have contributed to the successes and increase in diversity of this large and diverse group containing 11 tribes.
In short, Asteraceae have experienced several morphological changes, including the transition from a woody to herbaceous habit, from homogamous capitula with isomorphic florets to heterogamous capitula with differentiated florets, from a discoid capitulum to other types with zygomorphic florets in several large subfamilies, including radiate-like (Mutisioideae), ligulate (Cichorioideae I), and radiate (most Asteroideae and tribes in Cichorioideae II/Vernonioideae). The formation of paleate receptacles and variously modified pappuses increased the defense against herbivores and/or enhanced the dispersal of achenes. These morphological changes are associated with increases in diversity, suggesting that they might have played important roles in the elevated biodiversity of these groups.
CONCLUSIONS
We generated new transcriptome and genome datasets for 137 Asteraceae species and combined them with 106 other datasets (Zeng et al., 2014; Liu et al., 2015; Huang et al., 2016b) to provide a sample of 243 Asteraceae species, representing all 13 subfamilies and 41 of the 45 tribes, excluding only four small tribes with a total of nine species. These datasets were used to obtain >1,000 nuclear genes for phylogenetic analyses, molecular clock estimates, and analyses of diversification rate shifts, as well as >5,000 gene families for the phylogenomic detection of coincidental gene duplicates at multiple nodes in the Asteraceae phylogeny, providing strong evidence for 41 WGD events. The newly established nuclear Asteraceae phylogeny provides a well-supported resolution of most relationships among the sampled taxa, including the monophyly of eight of the 11 subfamilies containing more than one species and 30 of the 32 tribes with at least two species (Millerieae and Neurolaeneae are not shown to be monophyletic as currently circumscribed). In addition, this nuclear phylogeny is consistent with earlier plastid phylogenies for the relative placements of many subfamilies and tribes (see Panero and Crozier (2016) and references therein); however, several differences were found when comparing the plastid and nuclear phylogenies, some of which suggest possible hybrid origins of major lineages within the family. These include the phylogenetic positions of Cichorioideae, Pertyoideae, Stifftieae, Cichorieae, Hyalideae, Dicomeae, Anthemideae, Calenduleae, Gnaphalieae, all but two of the tribes of the Heliantheae alliance, and one genus each in Millerieae and Neurolaeneae. Three previously named subfamilies were found to be paraphyletic/polyphyletic, supporting the recognition of the tribe Dicomeae as the subfamily Dicomoideae, and the subfamily Vernonioideae for the clade containing the tribes Arctotideae, Liabeae, and Vernonieae. The two tribes of Wunderlichioideae are nested in different clades and may represent different subfamilies as well.
The well-resolved Asteraceae phylogeny proposed here provides the fundamental framework for further evolutionary analyses. Molecular clock estimates suggested that Asteraceae originated ~83 Mya in the late Cretaceous, that most early-divergent and small subfamilies diverged during the subsequent 10 My, and that further diversification, including the three largest subfamilies, occurred near the Cretaceous–Paleocene boundary (~66 Mya) (circles 2, 5–7 in Figure 3). Further diversification among most tribes was detected between the Eocene and the Paleocene (circle 4 in Figure 3). The results suggest that climate changes and the associated mass extinctions might have provided newly freed niches, which then were used by Asteraceae lineages that migrated to new locations and diversified, as supported by the proposed migration of the Asteraceae lineages (Mandel et al., 2019). Similar situations have also been suggested for Fabaceae, Solanaceae, and Poaceae (Vanneste et al., 2014b).
The Asteraceae topology at the tribal level and analyses of nuclear gene families from nearly all tribes allowed the detection of strong evidence for numerous WGDs using phylogenomic analyses. The detected WGDs are consistent with previous studies with smaller taxon samplings, including one shared by Asteraceae and its sister family Calyceraceae, one shared by the core Asteraceae (without Barnadesioideae and Famatinanthoideae), and one shared by the Heliantheae alliance (without/with Helenieae). In addition, a total of 32 candidate WGDs, some with minor support, were also proposed, including 19 in the largest subfamily, Asteroideae (Figure S35). Some candidate WGDs are associated with small subfamilies (WGD32 with Gymnarrhenoideae and WGD40 with Barnadesioideae), tribes (WGD21/WGD22 with Astereae, WGD23 with Gnaphalieae, and WGD37 with Wunderlichieae), or within a tribe (WGD29/WGD30 in Cichorioideae, and WGD33 in Cardueae). WGDs provide abundant genetic material for functional evolution, which likely contributed to morphological innovations, speciation, and adaptive radiation (Maere and Van de Peer, 2010; Arrigo and Barker, 2012). Such changes likely allowed organisms to take advantage of new ecological opportunities or cope with new environmental challenges, and might therefore have resulted in the expansion of biodiversity (Maere and Van de Peer, 2010; Schranz et al., 2012; Fawcett et al., 2013). Among the Asteraceae lineages, multiple pairs of sister clades (thus descendants of a common ancestor) exhibited great differences in both species number and the number of detected WGD events shared by the corresponding clade and/or its subgroups, including: (i) Barnadesioideae (92 species and two WGDs) versus the remaining Asteraceae (~25,900 species (assuming the total Asteraceae species number is ~26,000) and 37 WGDs); (ii) Famatinanthoideae (one species and no WGD) versus the core Asteraceae (~25,900 species and 37 WGDs); (iii) the Mutisioideae–Stifftioideae clade (~690 species and five WGDs) versus the remaining core Asteraceae (~25,200 species and 31 WGDs); (iv) a grade of four successive sister branches (each with one to 85 species and zero or one WGD) of the clade Carduoideae–Asteroideae (~25,000 species and 29 WGDs); and (v) Corymbioideae (nine species and no WGD) versus Asteroideae (>17,000 species and 23 WGDs). The Asteraceae WGDs might therefore have contributed to the increases in biodiversity associated with the highly successful major clades of the family.
Furthermore, the reconstruction of ancestral morphological characters supports key morphological changes during the history of Asteraceae. One of these changes is from a woody to herbaceous habit after the separation of Pertyoideae from the common ancestor of the three largest subfamilies. This habit change postdated the WGD shared by the core Asteraceae, but predated the upshift in diversification rate associated with the node of both Asteroideae and Cichorioideae. In addition, the capitulum type transitioned from discoid to radiate-like, ligulate, and radiate in the ancestors of Mutisioideae, Cichorioideae I, Asteroideae, and Cichorioideae II, respectively, which likely contributed to enhanced pollinator attraction by the functionally “flower-like” heads and increased reproductive success. It is worth noting that the smaller clades (e.g., Barnadesioideae, Famatinanthoideae, Wunderlichioideae II, Gochnatioideae, Hecastocleidoideae, Pertyoideae, and Corymbioideae) in most pairs of sister clades with vastly different sizes mentioned in the previous paragraph have discoid capitula, in contrast to the larger sister clades with many taxa having heads with zygomorphic outer florets (Figure S39). In addition, among the early-divergent subfamilies, the Mutisioideae–Stifftioideae clade (~690 spp.) contains many taxa with radiate-like heads, and is larger than the others with discoid heads (species numbers from 1 to <100). The fact that the groups with “flower-like” heads are much larger than their sister clades suggests that the independent innovations in the capitulum likely contributed to their reproductive and evolutionary successes. Previous analyses have revealed that the MADS-box and CYCLOIDEA2 (CYC2) floral regulatory genes duplicated since the origin of Asteraceae but before the divergence of Carduoideae I from Asteroideae and others, and that these genes play important roles in the development of the capitulum meristem and corolla in zygomorphic flowers (Chen et al., 2018; Elomaa et al., 2018). Furthermore, multiple innovations of the pappus, which can protect the achene and facilitate its dispersal, particularly those associated with the Heliantheae alliance, likely promoted the reproductive fitness and evolutionary success of the corresponding lineages. The association of these morphological changes with increases in diversity in these groups suggests that these morphological innovations are among the factors that could have contributed to the diversification of the family.
In summary, our analyses using a newly established Asteraceae nuclear phylogeny suggest environmental factors, including climate changes, and newly freed niches due to mass extinction, WGDs, and (subsequent) morphological innovations are some of the factors that likely contributed to the great biodiversity in Asteraceae, one of the two largest families of angiosperms. Some of the WGDs detected here likely provided new genetic materials for the evolution of novel functions, some of which might have supported morphological changes, such as the herbaceous habit for environmental adaptation (associated with the large clade containing all four of the largest subfamilies), the “flower-like” radiate head inflorescence with both peripheral ray flowers with large petals for pollinator attraction and central disc flowers for reproduction (associated with Asteroideae), and the pappus for seed dispersal. These morphological changes then facilitated the adaptation of the corresponding lineages during the periods following mass extinctions (at the Cretaceous–Paleocene and Eocene–Oligocene boundaries) and the promotion of the migration and expansion of Asteraceae from South America to other continents, with their descendants, especially those of the four largest subfamilies, eventually becoming distributed throughout most of the Earth′s terrestrial and wetland ecosystems.
MATERIALS AND METHODS
Transcriptome and genome sequencing, selection of nuclear genes and phylogenetic analyses
RNAs were isolated from leaves and/or floral buds of 122 species representing major Asteraceae lineages (see Tables S1, S2). DNAs were extracted from dry samples for 16 other species. Transcriptome and genome shotgun sequencing were performed by using Illumina HiSeq2500 or HiSeq3000. The RNA/DNA sequencing raw reads were assembled with Trinity v2013-11-10 (Grabherr et al., 2011) and SOAPdenovo 2.04-r240 (Xie et al., 2014), respectively. The raw reads of newly generated samples in this study were uploaded into the National Center for Biotechnology Information databases (The BioProject accession number: PRJNA636629). Three sets of low-copy nuclear genes were obtained from the 244 Asteraceae datasets and five outgroup species, resulting in 1,087 OGs for phylogenetic analyses (Figure S2; Table S9). Coalescence and supermatrix phylogenetic reconstruction analyses were performed using ASTRAL 4.10.6 (Mirarab et al., 2014) and RaxML v7.0.4 (Stamatakis, 2006), respectively.
Divergence time estimations and analyses for net diversification rate shifts
A maximum likelihood tree using gene set 11 (Figure S2) with additional samplings of Asterales (Tables S1, S2) was reconstructed for molecular clock analyses. The ages of Asteraceae lineages were estimated using the penalized likelihood and Bayesian methods implemented respectively in r8s v1.7.1 (Sanderson, 2003) and BEAST v2.4.3 (Bouckaert et al., 2014), using 14 or 15 fossils in five alternative calibration sets (see Supporting Information) mostly according to Smith et al. (2010), Magallón et al. (2015) and Panero and Crozier (2016). We used MEDUSA (Alfaro et al., 2009) implemented in the Geiger package (Harmon et al., 2008) of R (R Core Team, 2016) as well as the two algorithms (time-constant and time-variable algorithms) in BAMM v2.5 (Rabosky, 2014; Rabosky et al., 2014a, 2014b) (http://bamm-project.org/, last accessed November 6, 2016) to estimate the diversification rate shifts within Asteraceae.
Phylogenomic analyses for gene duplications as evidence for WGD
There were 6,059 OGs selected as a template from our previous study (Huang et al., 2016a) to search orthologous sequences of all species used in phylogenetic analyses, with the addition of five Asteraceae species (Pertya phylicoides, Liabellum palmeri, Macledium spinosum, Palafoxia arida, Xanthopappus subacaulis) which were sequenced more recently, to supplement the sampling of five corresponding tribes (Tables S1, S2). After selection of sequence quality, the remaining 5,282 OGs were used in subsequent reconstruction and mapping procedures. Gene trees were reconstructed using FastTree v2.1.4 (Price et al., 2009, 2010), and then the nodes of gene duplication in each gene tree were mapped and counted at the corresponding nodes in the species tree.
Reconstruction of ancestral states of morphological characters
Evolutionary histories of seven characters including habit, inflorescence, capitulum sexual differentiation, capitulum type, corollas in a capitulum, pappus and receptacular bract were estimated using Mesquite v3.10 (http://mesquiteproject.org) under a stochastic ML Markov k-state one-parameter model on a revised likelihood tree of combined 192 genes (set 11; Figure S2) with genus as unit. Details of the character matrix are provided in Table S6 (see Supporting Information for details).
ACKNOWLEDGEMENTS
We thank Beijing Botanical Garden, Bonn University Botanic Gardens, Guizhou Botanical Garden, Shanghai Chenshan Botanical Garden, South China Botanical Garden, Tel Aviv University Botanical Garden, UC Berkeley Botanical Garden, US Botanical Garden, Victoria Botanical Garden, and Zhihao Cheng, Zhixi Fu, Holly Forbes, Megan Hirst, Haihua Hu, Xiaojie Li, Fan Lu, Jinshuang Ma, Michal Monosov, Dunyan Tan, Huang-Lung Tsai, Weimin Ni, Li Song, Yaqiong Wang, Zehuan Wang, Ji Yang, Yushi Ye, Liping Zeng, Junwen Zhai, Guojin Zhang, Ning Zhang, and Shu Zhang for plant materials, Ji Qi for a script to map gene duplications, and Bao Nie, Wei Wang, and Guojin Zhang for technical assistance and discussion. This work was supported by funds from grants from the National Natural Science Foundation of China (Nos 31770242 and 31970224) and by funds from the Biology Department and the Huck Institutes of the Life Sciences at the Pennsylvania State University.
AUTHOR CONTRIBUTIONS
H.M. conceived of and designed the study. C.Z. selected the taxa and performed the phylogenetic analyses, and morphological character reconstruction. C.Z., M.L., H.M., Y.H., J.L.P., F.L., and T.G. contributed plant materials; C.Z. and Y.H. performed RNA and DNA isolation. M.L. performed preliminary phylogenetic analyses based on partial data. C.-H.H. designed and performed the dating and the diversification rate analyses and phylogenomics of whole genome duplication. C.Z., C.-H.H., and H.M. wrote the paper with contributions from all other authors. All authors read and approved the manuscript.






